
Farfantepenaeus duorarum pink shrimp associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.93
Get precalculated fractions of proteins
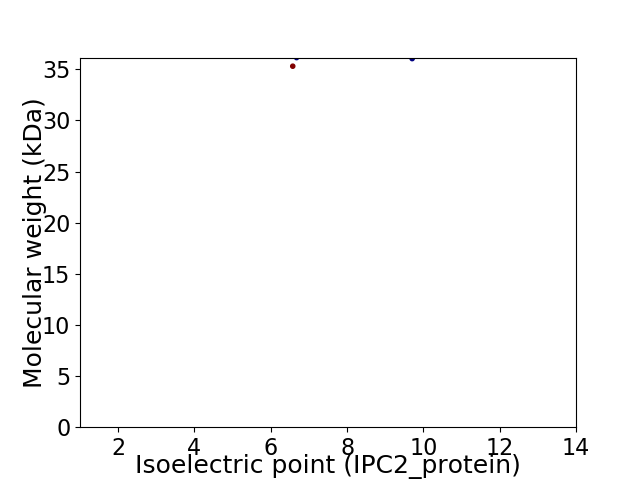
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RLN2|A0A0K1RLN2_9CIRC Putative replication initiation protein OS=Farfantepenaeus duorarum pink shrimp associated circular virus OX=1692248 PE=4 SV=1
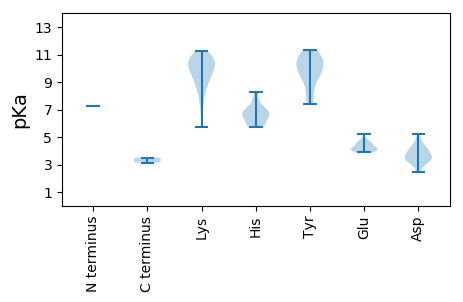
MM1 pKa = 7.26AQAKK5 pKa = 9.57RR6 pKa = 11.84WVFTLNNYY14 pKa = 9.94SGADD18 pKa = 3.62EE19 pKa = 4.12QLLEE23 pKa = 3.99QLSRR27 pKa = 11.84STDD30 pKa = 3.48VVTLIYY36 pKa = 10.42GRR38 pKa = 11.84EE39 pKa = 3.98VAPGTGTNHH48 pKa = 5.72LQGMIIFATRR58 pKa = 11.84KK59 pKa = 9.14RR60 pKa = 11.84LRR62 pKa = 11.84QIRR65 pKa = 11.84NYY67 pKa = 10.49PPFQRR72 pKa = 11.84AHH74 pKa = 6.55LEE76 pKa = 4.29VMRR79 pKa = 11.84STPARR84 pKa = 11.84ARR86 pKa = 11.84EE87 pKa = 3.92YY88 pKa = 10.55CIKK91 pKa = 11.0DD92 pKa = 3.13GDD94 pKa = 3.93FVEE97 pKa = 4.77YY98 pKa = 10.88GEE100 pKa = 4.21PVEE103 pKa = 4.45EE104 pKa = 3.99NRR106 pKa = 11.84QGRR109 pKa = 11.84RR110 pKa = 11.84NDD112 pKa = 2.8IHH114 pKa = 8.24DD115 pKa = 4.41IIAWLDD121 pKa = 3.46LFIEE125 pKa = 4.55EE126 pKa = 4.39NNRR129 pKa = 11.84PPTQEE134 pKa = 4.09EE135 pKa = 4.68VAQLQPIAMLRR146 pKa = 11.84YY147 pKa = 9.67RR148 pKa = 11.84NFMDD152 pKa = 4.89LARR155 pKa = 11.84LRR157 pKa = 11.84APQPQLIRR165 pKa = 11.84NGVARR170 pKa = 11.84EE171 pKa = 4.08GWQMDD176 pKa = 4.26LEE178 pKa = 4.57HH179 pKa = 7.54ALDD182 pKa = 4.3GDD184 pKa = 3.73APDD187 pKa = 3.48RR188 pKa = 11.84HH189 pKa = 6.94ILFYY193 pKa = 11.02VDD195 pKa = 3.33PAGGNGKK202 pKa = 7.29TWFQQYY208 pKa = 11.21LLTKK212 pKa = 8.89YY213 pKa = 9.63PRR215 pKa = 11.84RR216 pKa = 11.84VQILGTGNFADD227 pKa = 3.98CAYY230 pKa = 10.54AVDD233 pKa = 3.9EE234 pKa = 4.68SKK236 pKa = 11.17DD237 pKa = 3.13IFFFNVPRR245 pKa = 11.84GGMEE249 pKa = 4.03FFQYY253 pKa = 10.77RR254 pKa = 11.84LAEE257 pKa = 4.14SLKK260 pKa = 10.7DD261 pKa = 3.29RR262 pKa = 11.84LVFSTKK268 pKa = 9.66YY269 pKa = 10.1ASGMKK274 pKa = 9.4ILHH277 pKa = 6.14KK278 pKa = 10.34VPHH281 pKa = 5.78VVVFCNEE288 pKa = 3.89APDD291 pKa = 3.99MTKK294 pKa = 9.45LTADD298 pKa = 4.67RR299 pKa = 11.84IIIHH303 pKa = 6.78DD304 pKa = 4.16LL305 pKa = 3.5
MM1 pKa = 7.26AQAKK5 pKa = 9.57RR6 pKa = 11.84WVFTLNNYY14 pKa = 9.94SGADD18 pKa = 3.62EE19 pKa = 4.12QLLEE23 pKa = 3.99QLSRR27 pKa = 11.84STDD30 pKa = 3.48VVTLIYY36 pKa = 10.42GRR38 pKa = 11.84EE39 pKa = 3.98VAPGTGTNHH48 pKa = 5.72LQGMIIFATRR58 pKa = 11.84KK59 pKa = 9.14RR60 pKa = 11.84LRR62 pKa = 11.84QIRR65 pKa = 11.84NYY67 pKa = 10.49PPFQRR72 pKa = 11.84AHH74 pKa = 6.55LEE76 pKa = 4.29VMRR79 pKa = 11.84STPARR84 pKa = 11.84ARR86 pKa = 11.84EE87 pKa = 3.92YY88 pKa = 10.55CIKK91 pKa = 11.0DD92 pKa = 3.13GDD94 pKa = 3.93FVEE97 pKa = 4.77YY98 pKa = 10.88GEE100 pKa = 4.21PVEE103 pKa = 4.45EE104 pKa = 3.99NRR106 pKa = 11.84QGRR109 pKa = 11.84RR110 pKa = 11.84NDD112 pKa = 2.8IHH114 pKa = 8.24DD115 pKa = 4.41IIAWLDD121 pKa = 3.46LFIEE125 pKa = 4.55EE126 pKa = 4.39NNRR129 pKa = 11.84PPTQEE134 pKa = 4.09EE135 pKa = 4.68VAQLQPIAMLRR146 pKa = 11.84YY147 pKa = 9.67RR148 pKa = 11.84NFMDD152 pKa = 4.89LARR155 pKa = 11.84LRR157 pKa = 11.84APQPQLIRR165 pKa = 11.84NGVARR170 pKa = 11.84EE171 pKa = 4.08GWQMDD176 pKa = 4.26LEE178 pKa = 4.57HH179 pKa = 7.54ALDD182 pKa = 4.3GDD184 pKa = 3.73APDD187 pKa = 3.48RR188 pKa = 11.84HH189 pKa = 6.94ILFYY193 pKa = 11.02VDD195 pKa = 3.33PAGGNGKK202 pKa = 7.29TWFQQYY208 pKa = 11.21LLTKK212 pKa = 8.89YY213 pKa = 9.63PRR215 pKa = 11.84RR216 pKa = 11.84VQILGTGNFADD227 pKa = 3.98CAYY230 pKa = 10.54AVDD233 pKa = 3.9EE234 pKa = 4.68SKK236 pKa = 11.17DD237 pKa = 3.13IFFFNVPRR245 pKa = 11.84GGMEE249 pKa = 4.03FFQYY253 pKa = 10.77RR254 pKa = 11.84LAEE257 pKa = 4.14SLKK260 pKa = 10.7DD261 pKa = 3.29RR262 pKa = 11.84LVFSTKK268 pKa = 9.66YY269 pKa = 10.1ASGMKK274 pKa = 9.4ILHH277 pKa = 6.14KK278 pKa = 10.34VPHH281 pKa = 5.78VVVFCNEE288 pKa = 3.89APDD291 pKa = 3.99MTKK294 pKa = 9.45LTADD298 pKa = 4.67RR299 pKa = 11.84IIIHH303 pKa = 6.78DD304 pKa = 4.16LL305 pKa = 3.5
Molecular weight: 35.31 kDa
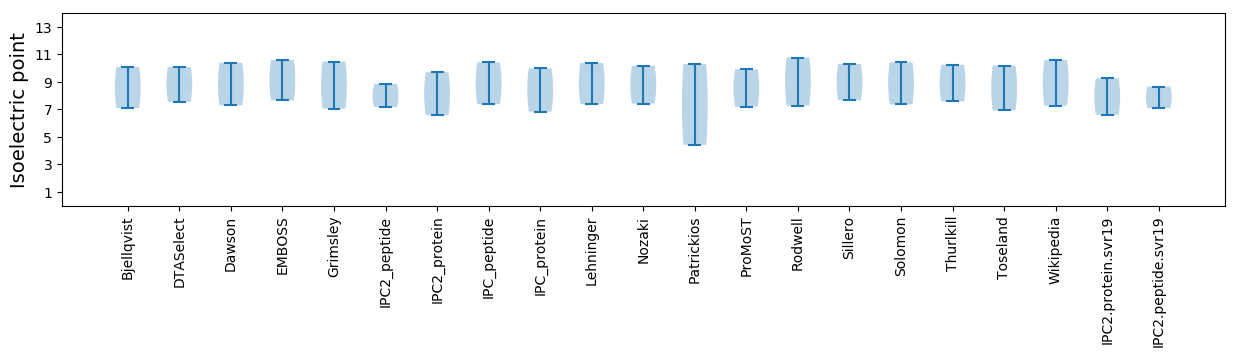
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RLN2|A0A0K1RLN2_9CIRC Putative replication initiation protein OS=Farfantepenaeus duorarum pink shrimp associated circular virus OX=1692248 PE=4 SV=1
MM1 pKa = 7.24SAIVPFARR9 pKa = 11.84YY10 pKa = 8.71AFPGARR16 pKa = 11.84RR17 pKa = 11.84AAAKK21 pKa = 10.53ALIRR25 pKa = 11.84TGKK28 pKa = 7.7ITPRR32 pKa = 11.84QYY34 pKa = 9.6MAAYY38 pKa = 9.24KK39 pKa = 9.98IGRR42 pKa = 11.84AAYY45 pKa = 8.39RR46 pKa = 11.84WRR48 pKa = 11.84GIFKK52 pKa = 10.45SKK54 pKa = 10.19GYY56 pKa = 9.77QRR58 pKa = 11.84SKK60 pKa = 10.6RR61 pKa = 11.84RR62 pKa = 11.84FVRR65 pKa = 11.84NVRR68 pKa = 11.84NVVRR72 pKa = 11.84NVRR75 pKa = 11.84SAQLKK80 pKa = 5.7TTHH83 pKa = 7.08LDD85 pKa = 3.49FASDD89 pKa = 3.46EE90 pKa = 4.21QFTNKK95 pKa = 9.71LNHH98 pKa = 6.0LHH100 pKa = 6.62HH101 pKa = 5.9VQPYY105 pKa = 8.17KK106 pKa = 10.63LVYY109 pKa = 10.57LGDD112 pKa = 4.22GDD114 pKa = 4.47NKK116 pKa = 10.29RR117 pKa = 11.84QSSTITCSGLRR128 pKa = 11.84VQFCFANTQPNGGQPQSHH146 pKa = 6.84LRR148 pKa = 11.84TDD150 pKa = 3.35YY151 pKa = 10.84DD152 pKa = 2.71IRR154 pKa = 11.84MVVLQEE160 pKa = 4.29KK161 pKa = 8.4PQPSYY166 pKa = 11.32VQDD169 pKa = 3.62DD170 pKa = 3.84VKK172 pKa = 11.22KK173 pKa = 10.45QFFSGRR179 pKa = 11.84GMAIDD184 pKa = 4.18YY185 pKa = 7.47GTIADD190 pKa = 5.2DD191 pKa = 3.63FTEE194 pKa = 5.22AYY196 pKa = 7.62PKK198 pKa = 10.5RR199 pKa = 11.84GIDD202 pKa = 3.72FQNLSGPMCQKK213 pKa = 8.65PTYY216 pKa = 9.05PINKK220 pKa = 8.97RR221 pKa = 11.84RR222 pKa = 11.84WQVLGNKK229 pKa = 10.13LIFLRR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84WDD238 pKa = 3.11GDD240 pKa = 3.25YY241 pKa = 11.32DD242 pKa = 3.82RR243 pKa = 11.84QAAGAWWVPLKK254 pKa = 10.73GKK256 pKa = 10.26KK257 pKa = 8.31LTYY260 pKa = 9.57TVTDD264 pKa = 3.25TGVEE268 pKa = 4.11YY269 pKa = 7.42TTPKK273 pKa = 9.95IYY275 pKa = 10.37FMYY278 pKa = 10.57YY279 pKa = 9.98IIPHH283 pKa = 6.69KK284 pKa = 10.66DD285 pKa = 2.46ITYY288 pKa = 10.01NLTHH292 pKa = 6.66ATTPVKK298 pKa = 10.8VDD300 pKa = 3.22IKK302 pKa = 9.68TIAYY306 pKa = 9.31FKK308 pKa = 9.09EE309 pKa = 4.51TNN311 pKa = 3.14
MM1 pKa = 7.24SAIVPFARR9 pKa = 11.84YY10 pKa = 8.71AFPGARR16 pKa = 11.84RR17 pKa = 11.84AAAKK21 pKa = 10.53ALIRR25 pKa = 11.84TGKK28 pKa = 7.7ITPRR32 pKa = 11.84QYY34 pKa = 9.6MAAYY38 pKa = 9.24KK39 pKa = 9.98IGRR42 pKa = 11.84AAYY45 pKa = 8.39RR46 pKa = 11.84WRR48 pKa = 11.84GIFKK52 pKa = 10.45SKK54 pKa = 10.19GYY56 pKa = 9.77QRR58 pKa = 11.84SKK60 pKa = 10.6RR61 pKa = 11.84RR62 pKa = 11.84FVRR65 pKa = 11.84NVRR68 pKa = 11.84NVVRR72 pKa = 11.84NVRR75 pKa = 11.84SAQLKK80 pKa = 5.7TTHH83 pKa = 7.08LDD85 pKa = 3.49FASDD89 pKa = 3.46EE90 pKa = 4.21QFTNKK95 pKa = 9.71LNHH98 pKa = 6.0LHH100 pKa = 6.62HH101 pKa = 5.9VQPYY105 pKa = 8.17KK106 pKa = 10.63LVYY109 pKa = 10.57LGDD112 pKa = 4.22GDD114 pKa = 4.47NKK116 pKa = 10.29RR117 pKa = 11.84QSSTITCSGLRR128 pKa = 11.84VQFCFANTQPNGGQPQSHH146 pKa = 6.84LRR148 pKa = 11.84TDD150 pKa = 3.35YY151 pKa = 10.84DD152 pKa = 2.71IRR154 pKa = 11.84MVVLQEE160 pKa = 4.29KK161 pKa = 8.4PQPSYY166 pKa = 11.32VQDD169 pKa = 3.62DD170 pKa = 3.84VKK172 pKa = 11.22KK173 pKa = 10.45QFFSGRR179 pKa = 11.84GMAIDD184 pKa = 4.18YY185 pKa = 7.47GTIADD190 pKa = 5.2DD191 pKa = 3.63FTEE194 pKa = 5.22AYY196 pKa = 7.62PKK198 pKa = 10.5RR199 pKa = 11.84GIDD202 pKa = 3.72FQNLSGPMCQKK213 pKa = 8.65PTYY216 pKa = 9.05PINKK220 pKa = 8.97RR221 pKa = 11.84RR222 pKa = 11.84WQVLGNKK229 pKa = 10.13LIFLRR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84WDD238 pKa = 3.11GDD240 pKa = 3.25YY241 pKa = 11.32DD242 pKa = 3.82RR243 pKa = 11.84QAAGAWWVPLKK254 pKa = 10.73GKK256 pKa = 10.26KK257 pKa = 8.31LTYY260 pKa = 9.57TVTDD264 pKa = 3.25TGVEE268 pKa = 4.11YY269 pKa = 7.42TTPKK273 pKa = 9.95IYY275 pKa = 10.37FMYY278 pKa = 10.57YY279 pKa = 9.98IIPHH283 pKa = 6.69KK284 pKa = 10.66DD285 pKa = 2.46ITYY288 pKa = 10.01NLTHH292 pKa = 6.66ATTPVKK298 pKa = 10.8VDD300 pKa = 3.22IKK302 pKa = 9.68TIAYY306 pKa = 9.31FKK308 pKa = 9.09EE309 pKa = 4.51TNN311 pKa = 3.14
Molecular weight: 36.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
616 |
305 |
311 |
308.0 |
35.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.63 ± 0.169 | 0.974 ± 0.007 |
6.169 ± 0.275 | 3.896 ± 1.65 |
5.032 ± 0.151 | 6.331 ± 0.072 |
2.435 ± 0.133 | 5.844 ± 0.041 |
5.844 ± 1.583 | 7.143 ± 1.209 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.435 ± 0.365 | 4.383 ± 0.146 |
5.195 ± 0.036 | 5.682 ± 0.076 |
8.929 ± 0.178 | 3.247 ± 0.441 |
6.169 ± 1.117 | 6.006 ± 0.074 |
1.461 ± 0.106 | 5.195 ± 0.891 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
