
Erysiphe necator mitovirus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.52
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I5ARF2|A0A2I5ARF2_9VIRU RNA-dependent RNA polymerase OS=Erysiphe necator mitovirus 3 OX=2052563 PE=4 SV=1
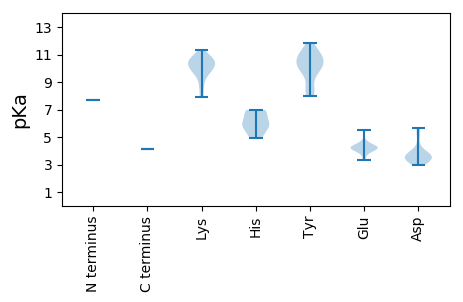
MM1 pKa = 7.71RR2 pKa = 11.84GVDD5 pKa = 3.37NTIQRR10 pKa = 11.84IKK12 pKa = 10.65DD13 pKa = 3.57SKK15 pKa = 10.72LHH17 pKa = 5.05ITRR20 pKa = 11.84YY21 pKa = 8.23MCGQALYY28 pKa = 10.94SSINPSVGINKK39 pKa = 9.7KK40 pKa = 10.17GLPIWIGTEE49 pKa = 3.92MNKK52 pKa = 10.28LADD55 pKa = 3.66GDD57 pKa = 3.64KK58 pKa = 9.98WEE60 pKa = 4.37KK61 pKa = 10.64RR62 pKa = 11.84LLLTLLSVSRR72 pKa = 11.84ALPGSQYY79 pKa = 10.5IPPLSSLSDD88 pKa = 3.09KK89 pKa = 9.73PTYY92 pKa = 10.47KK93 pKa = 9.98EE94 pKa = 4.32DD95 pKa = 4.17KK96 pKa = 8.04ISEE99 pKa = 4.43YY100 pKa = 10.2ISYY103 pKa = 10.44IPMVMQMMNIPSKK116 pKa = 11.02INVKK120 pKa = 8.59WRR122 pKa = 11.84QFHH125 pKa = 6.12LTTKK129 pKa = 10.33KK130 pKa = 10.77GPNGQAMVSSIRR142 pKa = 11.84DD143 pKa = 3.33AHH145 pKa = 6.97LLDD148 pKa = 5.66DD149 pKa = 5.65GMLKK153 pKa = 10.47DD154 pKa = 3.51IRR156 pKa = 11.84TVAGDD161 pKa = 3.7EE162 pKa = 4.03IIEE165 pKa = 4.49VINHH169 pKa = 5.45CRR171 pKa = 11.84EE172 pKa = 3.92LDD174 pKa = 3.69PEE176 pKa = 4.34SVEE179 pKa = 4.48VVAKK183 pKa = 10.96SMVKK187 pKa = 10.41PSTATLNEE195 pKa = 4.37DD196 pKa = 3.33KK197 pKa = 11.36GMDD200 pKa = 3.59KK201 pKa = 10.82PSTLLRR207 pKa = 11.84KK208 pKa = 10.06LSIVHH213 pKa = 5.77APEE216 pKa = 4.1RR217 pKa = 11.84KK218 pKa = 9.33SRR220 pKa = 11.84IIAILDD226 pKa = 3.25YY227 pKa = 8.25WTQSSLKK234 pKa = 8.84PLHH237 pKa = 6.83DD238 pKa = 5.31RR239 pKa = 11.84IFSILKK245 pKa = 10.73GIDD248 pKa = 3.03QDD250 pKa = 4.04YY251 pKa = 8.95TFNQQGVNKK260 pKa = 9.49RR261 pKa = 11.84LRR263 pKa = 11.84KK264 pKa = 10.0GPFYY268 pKa = 11.2SLDD271 pKa = 3.42LSSATDD277 pKa = 3.69RR278 pKa = 11.84FPLVFQQKK286 pKa = 7.92VVEE289 pKa = 4.16MLTCKK294 pKa = 10.04EE295 pKa = 4.19YY296 pKa = 11.08SEE298 pKa = 4.2AWSRR302 pKa = 11.84LLVNKK307 pKa = 9.51EE308 pKa = 4.65FYY310 pKa = 10.99VPWSDD315 pKa = 3.89SFINYY320 pKa = 9.69SCGQPMGAYY329 pKa = 9.92SSWSVFTLSHH339 pKa = 6.95HH340 pKa = 4.96ITVRR344 pKa = 11.84IAALMVGIPYY354 pKa = 9.77FDD356 pKa = 3.56QYY358 pKa = 11.83AILGDD363 pKa = 4.64DD364 pKa = 3.24IVIANSLVTKK374 pKa = 9.94KK375 pKa = 10.46YY376 pKa = 10.81KK377 pKa = 10.48EE378 pKa = 4.27IISDD382 pKa = 3.87LGVSISEE389 pKa = 4.33AKK391 pKa = 10.26SHH393 pKa = 6.09EE394 pKa = 4.23SSNTYY399 pKa = 10.11EE400 pKa = 4.2FAKK403 pKa = 9.59RR404 pKa = 11.84WYY406 pKa = 8.42VNSVEE411 pKa = 4.29VTGAQINAFMTKK423 pKa = 8.08STVWSTLINEE433 pKa = 4.35YY434 pKa = 10.97KK435 pKa = 10.23EE436 pKa = 4.59ICSKK440 pKa = 8.9WQIRR444 pKa = 11.84EE445 pKa = 3.94FEE447 pKa = 4.14TEE449 pKa = 3.31ARR451 pKa = 11.84IIYY454 pKa = 10.0DD455 pKa = 3.78LFLIFFPDD463 pKa = 2.97VSEE466 pKa = 4.3AFRR469 pKa = 11.84DD470 pKa = 3.57RR471 pKa = 11.84LVRR474 pKa = 11.84KK475 pKa = 9.7ARR477 pKa = 11.84IFLSLPWRR485 pKa = 11.84RR486 pKa = 11.84EE487 pKa = 3.78GTSASEE493 pKa = 4.1QILQFVLLTNASALGCFNSSTRR515 pKa = 11.84RR516 pKa = 11.84AKK518 pKa = 10.53EE519 pKa = 3.65FFVSSMAEE527 pKa = 3.46IKK529 pKa = 10.62ARR531 pKa = 11.84EE532 pKa = 4.09LNGGLIKK539 pKa = 10.25TNGLINDD546 pKa = 5.11LIKK549 pKa = 10.63KK550 pKa = 9.59FKK552 pKa = 10.7YY553 pKa = 10.64LLINYY558 pKa = 8.02KK559 pKa = 9.99GHH561 pKa = 5.78NHH563 pKa = 6.32HH564 pKa = 6.75EE565 pKa = 4.06ALKK568 pKa = 10.31AIPFVRR574 pKa = 11.84LILDD578 pKa = 3.31QRR580 pKa = 11.84VDD582 pKa = 2.95IMNSVDD588 pKa = 3.91LLQDD592 pKa = 3.63ASALTAEE599 pKa = 5.53DD600 pKa = 3.23IVLDD604 pKa = 4.1YY605 pKa = 10.44KK606 pKa = 10.97QKK608 pKa = 11.26APISTFDD615 pKa = 3.3PTKK618 pKa = 10.22ILTKK622 pKa = 9.59RR623 pKa = 11.84THH625 pKa = 5.78EE626 pKa = 4.8LILNRR631 pKa = 11.84DD632 pKa = 3.36ATIVNQYY639 pKa = 9.97SRR641 pKa = 11.84WIKK644 pKa = 10.2DD645 pKa = 3.35YY646 pKa = 11.53NVIKK650 pKa = 10.87QNVLNNSCDD659 pKa = 3.75EE660 pKa = 4.12NTEE663 pKa = 3.96RR664 pKa = 11.84AWARR668 pKa = 11.84KK669 pKa = 8.84CFRR672 pKa = 11.84TATIGAVMPGFPLL685 pKa = 4.1
MM1 pKa = 7.71RR2 pKa = 11.84GVDD5 pKa = 3.37NTIQRR10 pKa = 11.84IKK12 pKa = 10.65DD13 pKa = 3.57SKK15 pKa = 10.72LHH17 pKa = 5.05ITRR20 pKa = 11.84YY21 pKa = 8.23MCGQALYY28 pKa = 10.94SSINPSVGINKK39 pKa = 9.7KK40 pKa = 10.17GLPIWIGTEE49 pKa = 3.92MNKK52 pKa = 10.28LADD55 pKa = 3.66GDD57 pKa = 3.64KK58 pKa = 9.98WEE60 pKa = 4.37KK61 pKa = 10.64RR62 pKa = 11.84LLLTLLSVSRR72 pKa = 11.84ALPGSQYY79 pKa = 10.5IPPLSSLSDD88 pKa = 3.09KK89 pKa = 9.73PTYY92 pKa = 10.47KK93 pKa = 9.98EE94 pKa = 4.32DD95 pKa = 4.17KK96 pKa = 8.04ISEE99 pKa = 4.43YY100 pKa = 10.2ISYY103 pKa = 10.44IPMVMQMMNIPSKK116 pKa = 11.02INVKK120 pKa = 8.59WRR122 pKa = 11.84QFHH125 pKa = 6.12LTTKK129 pKa = 10.33KK130 pKa = 10.77GPNGQAMVSSIRR142 pKa = 11.84DD143 pKa = 3.33AHH145 pKa = 6.97LLDD148 pKa = 5.66DD149 pKa = 5.65GMLKK153 pKa = 10.47DD154 pKa = 3.51IRR156 pKa = 11.84TVAGDD161 pKa = 3.7EE162 pKa = 4.03IIEE165 pKa = 4.49VINHH169 pKa = 5.45CRR171 pKa = 11.84EE172 pKa = 3.92LDD174 pKa = 3.69PEE176 pKa = 4.34SVEE179 pKa = 4.48VVAKK183 pKa = 10.96SMVKK187 pKa = 10.41PSTATLNEE195 pKa = 4.37DD196 pKa = 3.33KK197 pKa = 11.36GMDD200 pKa = 3.59KK201 pKa = 10.82PSTLLRR207 pKa = 11.84KK208 pKa = 10.06LSIVHH213 pKa = 5.77APEE216 pKa = 4.1RR217 pKa = 11.84KK218 pKa = 9.33SRR220 pKa = 11.84IIAILDD226 pKa = 3.25YY227 pKa = 8.25WTQSSLKK234 pKa = 8.84PLHH237 pKa = 6.83DD238 pKa = 5.31RR239 pKa = 11.84IFSILKK245 pKa = 10.73GIDD248 pKa = 3.03QDD250 pKa = 4.04YY251 pKa = 8.95TFNQQGVNKK260 pKa = 9.49RR261 pKa = 11.84LRR263 pKa = 11.84KK264 pKa = 10.0GPFYY268 pKa = 11.2SLDD271 pKa = 3.42LSSATDD277 pKa = 3.69RR278 pKa = 11.84FPLVFQQKK286 pKa = 7.92VVEE289 pKa = 4.16MLTCKK294 pKa = 10.04EE295 pKa = 4.19YY296 pKa = 11.08SEE298 pKa = 4.2AWSRR302 pKa = 11.84LLVNKK307 pKa = 9.51EE308 pKa = 4.65FYY310 pKa = 10.99VPWSDD315 pKa = 3.89SFINYY320 pKa = 9.69SCGQPMGAYY329 pKa = 9.92SSWSVFTLSHH339 pKa = 6.95HH340 pKa = 4.96ITVRR344 pKa = 11.84IAALMVGIPYY354 pKa = 9.77FDD356 pKa = 3.56QYY358 pKa = 11.83AILGDD363 pKa = 4.64DD364 pKa = 3.24IVIANSLVTKK374 pKa = 9.94KK375 pKa = 10.46YY376 pKa = 10.81KK377 pKa = 10.48EE378 pKa = 4.27IISDD382 pKa = 3.87LGVSISEE389 pKa = 4.33AKK391 pKa = 10.26SHH393 pKa = 6.09EE394 pKa = 4.23SSNTYY399 pKa = 10.11EE400 pKa = 4.2FAKK403 pKa = 9.59RR404 pKa = 11.84WYY406 pKa = 8.42VNSVEE411 pKa = 4.29VTGAQINAFMTKK423 pKa = 8.08STVWSTLINEE433 pKa = 4.35YY434 pKa = 10.97KK435 pKa = 10.23EE436 pKa = 4.59ICSKK440 pKa = 8.9WQIRR444 pKa = 11.84EE445 pKa = 3.94FEE447 pKa = 4.14TEE449 pKa = 3.31ARR451 pKa = 11.84IIYY454 pKa = 10.0DD455 pKa = 3.78LFLIFFPDD463 pKa = 2.97VSEE466 pKa = 4.3AFRR469 pKa = 11.84DD470 pKa = 3.57RR471 pKa = 11.84LVRR474 pKa = 11.84KK475 pKa = 9.7ARR477 pKa = 11.84IFLSLPWRR485 pKa = 11.84RR486 pKa = 11.84EE487 pKa = 3.78GTSASEE493 pKa = 4.1QILQFVLLTNASALGCFNSSTRR515 pKa = 11.84RR516 pKa = 11.84AKK518 pKa = 10.53EE519 pKa = 3.65FFVSSMAEE527 pKa = 3.46IKK529 pKa = 10.62ARR531 pKa = 11.84EE532 pKa = 4.09LNGGLIKK539 pKa = 10.25TNGLINDD546 pKa = 5.11LIKK549 pKa = 10.63KK550 pKa = 9.59FKK552 pKa = 10.7YY553 pKa = 10.64LLINYY558 pKa = 8.02KK559 pKa = 9.99GHH561 pKa = 5.78NHH563 pKa = 6.32HH564 pKa = 6.75EE565 pKa = 4.06ALKK568 pKa = 10.31AIPFVRR574 pKa = 11.84LILDD578 pKa = 3.31QRR580 pKa = 11.84VDD582 pKa = 2.95IMNSVDD588 pKa = 3.91LLQDD592 pKa = 3.63ASALTAEE599 pKa = 5.53DD600 pKa = 3.23IVLDD604 pKa = 4.1YY605 pKa = 10.44KK606 pKa = 10.97QKK608 pKa = 11.26APISTFDD615 pKa = 3.3PTKK618 pKa = 10.22ILTKK622 pKa = 9.59RR623 pKa = 11.84THH625 pKa = 5.78EE626 pKa = 4.8LILNRR631 pKa = 11.84DD632 pKa = 3.36ATIVNQYY639 pKa = 9.97SRR641 pKa = 11.84WIKK644 pKa = 10.2DD645 pKa = 3.35YY646 pKa = 11.53NVIKK650 pKa = 10.87QNVLNNSCDD659 pKa = 3.75EE660 pKa = 4.12NTEE663 pKa = 3.96RR664 pKa = 11.84AWARR668 pKa = 11.84KK669 pKa = 8.84CFRR672 pKa = 11.84TATIGAVMPGFPLL685 pKa = 4.1
Molecular weight: 78.27 kDa
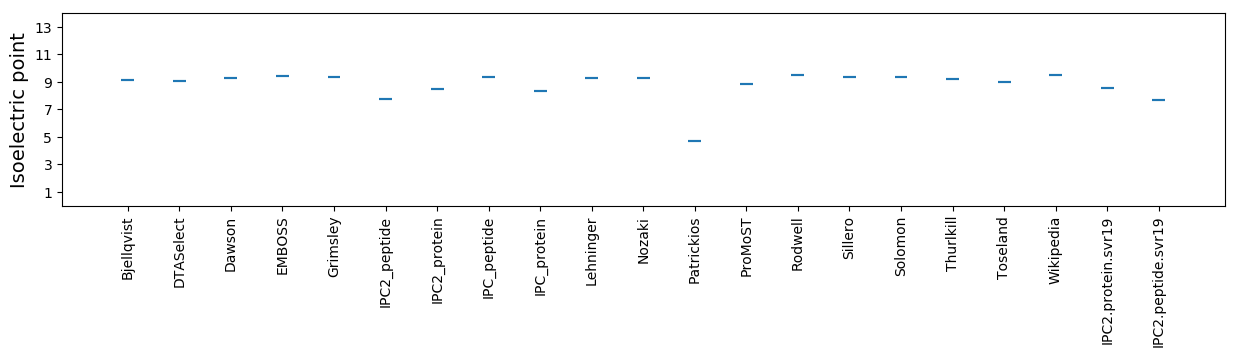
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I5ARF2|A0A2I5ARF2_9VIRU RNA-dependent RNA polymerase OS=Erysiphe necator mitovirus 3 OX=2052563 PE=4 SV=1
MM1 pKa = 7.71RR2 pKa = 11.84GVDD5 pKa = 3.37NTIQRR10 pKa = 11.84IKK12 pKa = 10.65DD13 pKa = 3.57SKK15 pKa = 10.72LHH17 pKa = 5.05ITRR20 pKa = 11.84YY21 pKa = 8.23MCGQALYY28 pKa = 10.94SSINPSVGINKK39 pKa = 9.7KK40 pKa = 10.17GLPIWIGTEE49 pKa = 3.92MNKK52 pKa = 10.28LADD55 pKa = 3.66GDD57 pKa = 3.64KK58 pKa = 9.98WEE60 pKa = 4.37KK61 pKa = 10.64RR62 pKa = 11.84LLLTLLSVSRR72 pKa = 11.84ALPGSQYY79 pKa = 10.5IPPLSSLSDD88 pKa = 3.09KK89 pKa = 9.73PTYY92 pKa = 10.47KK93 pKa = 9.98EE94 pKa = 4.32DD95 pKa = 4.17KK96 pKa = 8.04ISEE99 pKa = 4.43YY100 pKa = 10.2ISYY103 pKa = 10.44IPMVMQMMNIPSKK116 pKa = 11.02INVKK120 pKa = 8.59WRR122 pKa = 11.84QFHH125 pKa = 6.12LTTKK129 pKa = 10.33KK130 pKa = 10.77GPNGQAMVSSIRR142 pKa = 11.84DD143 pKa = 3.33AHH145 pKa = 6.97LLDD148 pKa = 5.66DD149 pKa = 5.65GMLKK153 pKa = 10.47DD154 pKa = 3.51IRR156 pKa = 11.84TVAGDD161 pKa = 3.7EE162 pKa = 4.03IIEE165 pKa = 4.49VINHH169 pKa = 5.45CRR171 pKa = 11.84EE172 pKa = 3.92LDD174 pKa = 3.69PEE176 pKa = 4.34SVEE179 pKa = 4.48VVAKK183 pKa = 10.96SMVKK187 pKa = 10.41PSTATLNEE195 pKa = 4.37DD196 pKa = 3.33KK197 pKa = 11.36GMDD200 pKa = 3.59KK201 pKa = 10.82PSTLLRR207 pKa = 11.84KK208 pKa = 10.06LSIVHH213 pKa = 5.77APEE216 pKa = 4.1RR217 pKa = 11.84KK218 pKa = 9.33SRR220 pKa = 11.84IIAILDD226 pKa = 3.25YY227 pKa = 8.25WTQSSLKK234 pKa = 8.84PLHH237 pKa = 6.83DD238 pKa = 5.31RR239 pKa = 11.84IFSILKK245 pKa = 10.73GIDD248 pKa = 3.03QDD250 pKa = 4.04YY251 pKa = 8.95TFNQQGVNKK260 pKa = 9.49RR261 pKa = 11.84LRR263 pKa = 11.84KK264 pKa = 10.0GPFYY268 pKa = 11.2SLDD271 pKa = 3.42LSSATDD277 pKa = 3.69RR278 pKa = 11.84FPLVFQQKK286 pKa = 7.92VVEE289 pKa = 4.16MLTCKK294 pKa = 10.04EE295 pKa = 4.19YY296 pKa = 11.08SEE298 pKa = 4.2AWSRR302 pKa = 11.84LLVNKK307 pKa = 9.51EE308 pKa = 4.65FYY310 pKa = 10.99VPWSDD315 pKa = 3.89SFINYY320 pKa = 9.69SCGQPMGAYY329 pKa = 9.92SSWSVFTLSHH339 pKa = 6.95HH340 pKa = 4.96ITVRR344 pKa = 11.84IAALMVGIPYY354 pKa = 9.77FDD356 pKa = 3.56QYY358 pKa = 11.83AILGDD363 pKa = 4.64DD364 pKa = 3.24IVIANSLVTKK374 pKa = 9.94KK375 pKa = 10.46YY376 pKa = 10.81KK377 pKa = 10.48EE378 pKa = 4.27IISDD382 pKa = 3.87LGVSISEE389 pKa = 4.33AKK391 pKa = 10.26SHH393 pKa = 6.09EE394 pKa = 4.23SSNTYY399 pKa = 10.11EE400 pKa = 4.2FAKK403 pKa = 9.59RR404 pKa = 11.84WYY406 pKa = 8.42VNSVEE411 pKa = 4.29VTGAQINAFMTKK423 pKa = 8.08STVWSTLINEE433 pKa = 4.35YY434 pKa = 10.97KK435 pKa = 10.23EE436 pKa = 4.59ICSKK440 pKa = 8.9WQIRR444 pKa = 11.84EE445 pKa = 3.94FEE447 pKa = 4.14TEE449 pKa = 3.31ARR451 pKa = 11.84IIYY454 pKa = 10.0DD455 pKa = 3.78LFLIFFPDD463 pKa = 2.97VSEE466 pKa = 4.3AFRR469 pKa = 11.84DD470 pKa = 3.57RR471 pKa = 11.84LVRR474 pKa = 11.84KK475 pKa = 9.7ARR477 pKa = 11.84IFLSLPWRR485 pKa = 11.84RR486 pKa = 11.84EE487 pKa = 3.78GTSASEE493 pKa = 4.1QILQFVLLTNASALGCFNSSTRR515 pKa = 11.84RR516 pKa = 11.84AKK518 pKa = 10.53EE519 pKa = 3.65FFVSSMAEE527 pKa = 3.46IKK529 pKa = 10.62ARR531 pKa = 11.84EE532 pKa = 4.09LNGGLIKK539 pKa = 10.25TNGLINDD546 pKa = 5.11LIKK549 pKa = 10.63KK550 pKa = 9.59FKK552 pKa = 10.7YY553 pKa = 10.64LLINYY558 pKa = 8.02KK559 pKa = 9.99GHH561 pKa = 5.78NHH563 pKa = 6.32HH564 pKa = 6.75EE565 pKa = 4.06ALKK568 pKa = 10.31AIPFVRR574 pKa = 11.84LILDD578 pKa = 3.31QRR580 pKa = 11.84VDD582 pKa = 2.95IMNSVDD588 pKa = 3.91LLQDD592 pKa = 3.63ASALTAEE599 pKa = 5.53DD600 pKa = 3.23IVLDD604 pKa = 4.1YY605 pKa = 10.44KK606 pKa = 10.97QKK608 pKa = 11.26APISTFDD615 pKa = 3.3PTKK618 pKa = 10.22ILTKK622 pKa = 9.59RR623 pKa = 11.84THH625 pKa = 5.78EE626 pKa = 4.8LILNRR631 pKa = 11.84DD632 pKa = 3.36ATIVNQYY639 pKa = 9.97SRR641 pKa = 11.84WIKK644 pKa = 10.2DD645 pKa = 3.35YY646 pKa = 11.53NVIKK650 pKa = 10.87QNVLNNSCDD659 pKa = 3.75EE660 pKa = 4.12NTEE663 pKa = 3.96RR664 pKa = 11.84AWARR668 pKa = 11.84KK669 pKa = 8.84CFRR672 pKa = 11.84TATIGAVMPGFPLL685 pKa = 4.1
MM1 pKa = 7.71RR2 pKa = 11.84GVDD5 pKa = 3.37NTIQRR10 pKa = 11.84IKK12 pKa = 10.65DD13 pKa = 3.57SKK15 pKa = 10.72LHH17 pKa = 5.05ITRR20 pKa = 11.84YY21 pKa = 8.23MCGQALYY28 pKa = 10.94SSINPSVGINKK39 pKa = 9.7KK40 pKa = 10.17GLPIWIGTEE49 pKa = 3.92MNKK52 pKa = 10.28LADD55 pKa = 3.66GDD57 pKa = 3.64KK58 pKa = 9.98WEE60 pKa = 4.37KK61 pKa = 10.64RR62 pKa = 11.84LLLTLLSVSRR72 pKa = 11.84ALPGSQYY79 pKa = 10.5IPPLSSLSDD88 pKa = 3.09KK89 pKa = 9.73PTYY92 pKa = 10.47KK93 pKa = 9.98EE94 pKa = 4.32DD95 pKa = 4.17KK96 pKa = 8.04ISEE99 pKa = 4.43YY100 pKa = 10.2ISYY103 pKa = 10.44IPMVMQMMNIPSKK116 pKa = 11.02INVKK120 pKa = 8.59WRR122 pKa = 11.84QFHH125 pKa = 6.12LTTKK129 pKa = 10.33KK130 pKa = 10.77GPNGQAMVSSIRR142 pKa = 11.84DD143 pKa = 3.33AHH145 pKa = 6.97LLDD148 pKa = 5.66DD149 pKa = 5.65GMLKK153 pKa = 10.47DD154 pKa = 3.51IRR156 pKa = 11.84TVAGDD161 pKa = 3.7EE162 pKa = 4.03IIEE165 pKa = 4.49VINHH169 pKa = 5.45CRR171 pKa = 11.84EE172 pKa = 3.92LDD174 pKa = 3.69PEE176 pKa = 4.34SVEE179 pKa = 4.48VVAKK183 pKa = 10.96SMVKK187 pKa = 10.41PSTATLNEE195 pKa = 4.37DD196 pKa = 3.33KK197 pKa = 11.36GMDD200 pKa = 3.59KK201 pKa = 10.82PSTLLRR207 pKa = 11.84KK208 pKa = 10.06LSIVHH213 pKa = 5.77APEE216 pKa = 4.1RR217 pKa = 11.84KK218 pKa = 9.33SRR220 pKa = 11.84IIAILDD226 pKa = 3.25YY227 pKa = 8.25WTQSSLKK234 pKa = 8.84PLHH237 pKa = 6.83DD238 pKa = 5.31RR239 pKa = 11.84IFSILKK245 pKa = 10.73GIDD248 pKa = 3.03QDD250 pKa = 4.04YY251 pKa = 8.95TFNQQGVNKK260 pKa = 9.49RR261 pKa = 11.84LRR263 pKa = 11.84KK264 pKa = 10.0GPFYY268 pKa = 11.2SLDD271 pKa = 3.42LSSATDD277 pKa = 3.69RR278 pKa = 11.84FPLVFQQKK286 pKa = 7.92VVEE289 pKa = 4.16MLTCKK294 pKa = 10.04EE295 pKa = 4.19YY296 pKa = 11.08SEE298 pKa = 4.2AWSRR302 pKa = 11.84LLVNKK307 pKa = 9.51EE308 pKa = 4.65FYY310 pKa = 10.99VPWSDD315 pKa = 3.89SFINYY320 pKa = 9.69SCGQPMGAYY329 pKa = 9.92SSWSVFTLSHH339 pKa = 6.95HH340 pKa = 4.96ITVRR344 pKa = 11.84IAALMVGIPYY354 pKa = 9.77FDD356 pKa = 3.56QYY358 pKa = 11.83AILGDD363 pKa = 4.64DD364 pKa = 3.24IVIANSLVTKK374 pKa = 9.94KK375 pKa = 10.46YY376 pKa = 10.81KK377 pKa = 10.48EE378 pKa = 4.27IISDD382 pKa = 3.87LGVSISEE389 pKa = 4.33AKK391 pKa = 10.26SHH393 pKa = 6.09EE394 pKa = 4.23SSNTYY399 pKa = 10.11EE400 pKa = 4.2FAKK403 pKa = 9.59RR404 pKa = 11.84WYY406 pKa = 8.42VNSVEE411 pKa = 4.29VTGAQINAFMTKK423 pKa = 8.08STVWSTLINEE433 pKa = 4.35YY434 pKa = 10.97KK435 pKa = 10.23EE436 pKa = 4.59ICSKK440 pKa = 8.9WQIRR444 pKa = 11.84EE445 pKa = 3.94FEE447 pKa = 4.14TEE449 pKa = 3.31ARR451 pKa = 11.84IIYY454 pKa = 10.0DD455 pKa = 3.78LFLIFFPDD463 pKa = 2.97VSEE466 pKa = 4.3AFRR469 pKa = 11.84DD470 pKa = 3.57RR471 pKa = 11.84LVRR474 pKa = 11.84KK475 pKa = 9.7ARR477 pKa = 11.84IFLSLPWRR485 pKa = 11.84RR486 pKa = 11.84EE487 pKa = 3.78GTSASEE493 pKa = 4.1QILQFVLLTNASALGCFNSSTRR515 pKa = 11.84RR516 pKa = 11.84AKK518 pKa = 10.53EE519 pKa = 3.65FFVSSMAEE527 pKa = 3.46IKK529 pKa = 10.62ARR531 pKa = 11.84EE532 pKa = 4.09LNGGLIKK539 pKa = 10.25TNGLINDD546 pKa = 5.11LIKK549 pKa = 10.63KK550 pKa = 9.59FKK552 pKa = 10.7YY553 pKa = 10.64LLINYY558 pKa = 8.02KK559 pKa = 9.99GHH561 pKa = 5.78NHH563 pKa = 6.32HH564 pKa = 6.75EE565 pKa = 4.06ALKK568 pKa = 10.31AIPFVRR574 pKa = 11.84LILDD578 pKa = 3.31QRR580 pKa = 11.84VDD582 pKa = 2.95IMNSVDD588 pKa = 3.91LLQDD592 pKa = 3.63ASALTAEE599 pKa = 5.53DD600 pKa = 3.23IVLDD604 pKa = 4.1YY605 pKa = 10.44KK606 pKa = 10.97QKK608 pKa = 11.26APISTFDD615 pKa = 3.3PTKK618 pKa = 10.22ILTKK622 pKa = 9.59RR623 pKa = 11.84THH625 pKa = 5.78EE626 pKa = 4.8LILNRR631 pKa = 11.84DD632 pKa = 3.36ATIVNQYY639 pKa = 9.97SRR641 pKa = 11.84WIKK644 pKa = 10.2DD645 pKa = 3.35YY646 pKa = 11.53NVIKK650 pKa = 10.87QNVLNNSCDD659 pKa = 3.75EE660 pKa = 4.12NTEE663 pKa = 3.96RR664 pKa = 11.84AWARR668 pKa = 11.84KK669 pKa = 8.84CFRR672 pKa = 11.84TATIGAVMPGFPLL685 pKa = 4.1
Molecular weight: 78.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
685 |
685 |
685 |
685.0 |
78.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.985 ± 0.0 | 1.168 ± 0.0 |
5.693 ± 0.0 | 5.255 ± 0.0 |
3.942 ± 0.0 | 4.234 ± 0.0 |
1.898 ± 0.0 | 8.759 ± 0.0 |
7.591 ± 0.0 | 9.489 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.628 ± 0.0 | 4.818 ± 0.0 |
3.796 ± 0.0 | 3.358 ± 0.0 |
5.547 ± 0.0 | 8.905 ± 0.0 |
5.401 ± 0.0 | 5.985 ± 0.0 |
1.898 ± 0.0 | 3.65 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
