
Paenibacillus anaericanus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Paenibacillus
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
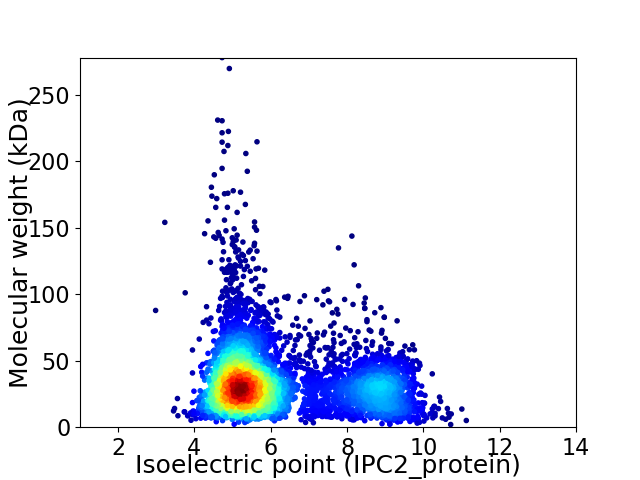
Virtual 2D-PAGE plot for 5415 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
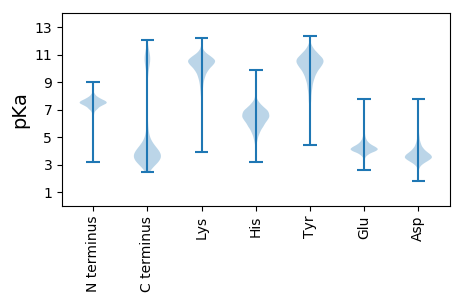
Protein with the lowest isoelectric point:
>tr|A0A3S1BG44|A0A3S1BG44_9BACL Uncharacterized protein OS=Paenibacillus anaericanus OX=170367 GN=EJP82_26755 PE=4 SV=1
MM1 pKa = 7.32SAIDD5 pKa = 3.53AQQRR9 pKa = 11.84KK10 pKa = 8.71VMDD13 pKa = 3.9SYY15 pKa = 12.09GKK17 pKa = 9.86IPLTFVEE24 pKa = 4.79NCGQTQANIRR34 pKa = 11.84YY35 pKa = 5.35YY36 pKa = 9.65TKK38 pKa = 9.9RR39 pKa = 11.84LGYY42 pKa = 10.09GVYY45 pKa = 8.33FTPNEE50 pKa = 3.63AAFFFRR56 pKa = 11.84DD57 pKa = 3.17NSRR60 pKa = 11.84QEE62 pKa = 4.06NVEE65 pKa = 3.71QSLFLQFPNANPEE78 pKa = 3.99VKK80 pKa = 10.73LEE82 pKa = 3.87GRR84 pKa = 11.84VKK86 pKa = 10.72GSEE89 pKa = 3.96KK90 pKa = 10.88VNFLIGNDD98 pKa = 3.39PTKK101 pKa = 10.03WHH103 pKa = 6.89TGLSTYY109 pKa = 10.16QEE111 pKa = 3.99VVYY114 pKa = 10.7RR115 pKa = 11.84EE116 pKa = 4.01LWSGVDD122 pKa = 3.34LVFYY126 pKa = 10.56EE127 pKa = 4.91GEE129 pKa = 4.25GKK131 pKa = 10.48LKK133 pKa = 10.64YY134 pKa = 10.22DD135 pKa = 5.23VILQPHH141 pKa = 6.69AKK143 pKa = 10.23VNDD146 pKa = 3.56VQLTYY151 pKa = 10.56HH152 pKa = 6.42GSDD155 pKa = 3.29YY156 pKa = 11.32LSLNEE161 pKa = 4.36EE162 pKa = 4.11GDD164 pKa = 4.2LLVHH168 pKa = 6.48TSLGVWTEE176 pKa = 3.98KK177 pKa = 10.87QPVSYY182 pKa = 11.02QEE184 pKa = 3.95INGKK188 pKa = 9.51KK189 pKa = 9.74VFVEE193 pKa = 4.1SRR195 pKa = 11.84FVLKK199 pKa = 9.54EE200 pKa = 3.82TDD202 pKa = 3.39QGDD205 pKa = 3.63QVYY208 pKa = 9.46GFEE211 pKa = 4.58MGQDD215 pKa = 3.67YY216 pKa = 11.35NPDD219 pKa = 3.44YY220 pKa = 11.31VLVIDD225 pKa = 5.67PILMYY230 pKa = 10.16STFLGGSVGPDD241 pKa = 2.94AGQGIAVDD249 pKa = 3.95TAGNAYY255 pKa = 8.37VTGFTAGSFPTTSGAYY271 pKa = 6.66QTIFGGDD278 pKa = 3.07SDD280 pKa = 4.96VFITKK285 pKa = 9.3MNSTGRR291 pKa = 11.84APIYY295 pKa = 9.31STYY298 pKa = 10.82LGGSGTDD305 pKa = 3.12VGQGITVDD313 pKa = 3.69TSGNAYY319 pKa = 7.83VTGYY323 pKa = 9.2TNSNDD328 pKa = 3.09FPTTVGAFQISLGGVGATNAFVTKK352 pKa = 10.54LNATGTLALYY362 pKa = 8.51STYY365 pKa = 10.92LGGDD369 pKa = 3.5TVDD372 pKa = 2.95IGYY375 pKa = 10.35GIAIDD380 pKa = 3.88ASGNAYY386 pKa = 8.38VTGSTTSGNYY396 pKa = 9.19PVTAGIFQGTLGGPGATNAFVTKK419 pKa = 10.61LNATGTAPLVYY430 pKa = 9.14STYY433 pKa = 11.04LGGTVSDD440 pKa = 3.83VGQGIAVDD448 pKa = 4.0ALGNAYY454 pKa = 8.32VTGSTTSTDD463 pKa = 3.87FPMVAGSVQTTYY475 pKa = 11.03GGSGDD480 pKa = 4.08AFVTKK485 pKa = 10.67LNATATAPLIYY496 pKa = 9.79STFLGGNGLDD506 pKa = 2.93IGYY509 pKa = 10.15SIGIDD514 pKa = 3.21AAGNAYY520 pKa = 9.54IAGFTDD526 pKa = 3.38SANYY530 pKa = 8.85PVEE533 pKa = 4.55GQIQPYY539 pKa = 9.73GGAGDD544 pKa = 4.3AMVSKK549 pKa = 10.51LNPAGTGLVYY559 pKa = 9.82STYY562 pKa = 10.73LGGIARR568 pKa = 11.84DD569 pKa = 3.44EE570 pKa = 3.97ARR572 pKa = 11.84AIVVDD577 pKa = 3.88PAGNAYY583 pKa = 8.82LTGNTEE589 pKa = 3.84SSDD592 pKa = 3.71YY593 pKa = 9.47PLSVGAIQTGLLGVRR608 pKa = 11.84DD609 pKa = 3.8VFITLFNTAGNALVYY624 pKa = 9.45STYY627 pKa = 10.98LGGTGVEE634 pKa = 4.2EE635 pKa = 4.37GLGIAVDD642 pKa = 4.24SNFNLYY648 pKa = 8.15VTGSTTSLNFPITPGAFQTSLPGLPGSGFVTKK680 pKa = 10.31IGTPIQLLGEE690 pKa = 4.34TGATGDD696 pKa = 4.18PGVTGPTGPTGDD708 pKa = 3.53TGATGEE714 pKa = 4.28TGATGEE720 pKa = 4.28TGATGEE726 pKa = 4.37TGATGDD732 pKa = 3.54TGATGEE738 pKa = 4.38TGATGDD744 pKa = 3.54TGATGEE750 pKa = 4.38TGATGDD756 pKa = 3.73TGPTGGTGATGDD768 pKa = 3.45TGATGEE774 pKa = 4.38TGATGDD780 pKa = 3.73TGPTGGTGATGDD792 pKa = 3.49TGATGGTGATGGTGATGEE810 pKa = 4.28TGATGDD816 pKa = 3.67TGATGDD822 pKa = 3.89TGATGATGEE831 pKa = 4.41TGATGDD837 pKa = 3.54TGATGEE843 pKa = 4.38TGATGDD849 pKa = 3.4TGAIGGTGATGDD861 pKa = 3.58TGATGDD867 pKa = 3.63TGATGEE873 pKa = 4.25TGATGGTGAAGEE885 pKa = 4.27TGATGEE891 pKa = 4.37TGATGDD897 pKa = 3.54TGATGEE903 pKa = 4.38TGATGDD909 pKa = 3.54TGATGEE915 pKa = 4.38TGATGDD921 pKa = 3.73TGPTGGTGATGDD933 pKa = 3.49TGATGGTGATGGTGATGEE951 pKa = 4.28TGATGDD957 pKa = 3.54TGATGEE963 pKa = 4.28TGEE966 pKa = 4.42TGATGEE972 pKa = 4.37TGATGDD978 pKa = 3.54TGATGEE984 pKa = 4.38TGATGDD990 pKa = 3.4TGAIGGTGATGDD1002 pKa = 3.58TGATGDD1008 pKa = 3.63TGATGEE1014 pKa = 4.25TGATGGTGAAGEE1026 pKa = 4.27TGATGEE1032 pKa = 4.41TGATGG1037 pKa = 3.17
MM1 pKa = 7.32SAIDD5 pKa = 3.53AQQRR9 pKa = 11.84KK10 pKa = 8.71VMDD13 pKa = 3.9SYY15 pKa = 12.09GKK17 pKa = 9.86IPLTFVEE24 pKa = 4.79NCGQTQANIRR34 pKa = 11.84YY35 pKa = 5.35YY36 pKa = 9.65TKK38 pKa = 9.9RR39 pKa = 11.84LGYY42 pKa = 10.09GVYY45 pKa = 8.33FTPNEE50 pKa = 3.63AAFFFRR56 pKa = 11.84DD57 pKa = 3.17NSRR60 pKa = 11.84QEE62 pKa = 4.06NVEE65 pKa = 3.71QSLFLQFPNANPEE78 pKa = 3.99VKK80 pKa = 10.73LEE82 pKa = 3.87GRR84 pKa = 11.84VKK86 pKa = 10.72GSEE89 pKa = 3.96KK90 pKa = 10.88VNFLIGNDD98 pKa = 3.39PTKK101 pKa = 10.03WHH103 pKa = 6.89TGLSTYY109 pKa = 10.16QEE111 pKa = 3.99VVYY114 pKa = 10.7RR115 pKa = 11.84EE116 pKa = 4.01LWSGVDD122 pKa = 3.34LVFYY126 pKa = 10.56EE127 pKa = 4.91GEE129 pKa = 4.25GKK131 pKa = 10.48LKK133 pKa = 10.64YY134 pKa = 10.22DD135 pKa = 5.23VILQPHH141 pKa = 6.69AKK143 pKa = 10.23VNDD146 pKa = 3.56VQLTYY151 pKa = 10.56HH152 pKa = 6.42GSDD155 pKa = 3.29YY156 pKa = 11.32LSLNEE161 pKa = 4.36EE162 pKa = 4.11GDD164 pKa = 4.2LLVHH168 pKa = 6.48TSLGVWTEE176 pKa = 3.98KK177 pKa = 10.87QPVSYY182 pKa = 11.02QEE184 pKa = 3.95INGKK188 pKa = 9.51KK189 pKa = 9.74VFVEE193 pKa = 4.1SRR195 pKa = 11.84FVLKK199 pKa = 9.54EE200 pKa = 3.82TDD202 pKa = 3.39QGDD205 pKa = 3.63QVYY208 pKa = 9.46GFEE211 pKa = 4.58MGQDD215 pKa = 3.67YY216 pKa = 11.35NPDD219 pKa = 3.44YY220 pKa = 11.31VLVIDD225 pKa = 5.67PILMYY230 pKa = 10.16STFLGGSVGPDD241 pKa = 2.94AGQGIAVDD249 pKa = 3.95TAGNAYY255 pKa = 8.37VTGFTAGSFPTTSGAYY271 pKa = 6.66QTIFGGDD278 pKa = 3.07SDD280 pKa = 4.96VFITKK285 pKa = 9.3MNSTGRR291 pKa = 11.84APIYY295 pKa = 9.31STYY298 pKa = 10.82LGGSGTDD305 pKa = 3.12VGQGITVDD313 pKa = 3.69TSGNAYY319 pKa = 7.83VTGYY323 pKa = 9.2TNSNDD328 pKa = 3.09FPTTVGAFQISLGGVGATNAFVTKK352 pKa = 10.54LNATGTLALYY362 pKa = 8.51STYY365 pKa = 10.92LGGDD369 pKa = 3.5TVDD372 pKa = 2.95IGYY375 pKa = 10.35GIAIDD380 pKa = 3.88ASGNAYY386 pKa = 8.38VTGSTTSGNYY396 pKa = 9.19PVTAGIFQGTLGGPGATNAFVTKK419 pKa = 10.61LNATGTAPLVYY430 pKa = 9.14STYY433 pKa = 11.04LGGTVSDD440 pKa = 3.83VGQGIAVDD448 pKa = 4.0ALGNAYY454 pKa = 8.32VTGSTTSTDD463 pKa = 3.87FPMVAGSVQTTYY475 pKa = 11.03GGSGDD480 pKa = 4.08AFVTKK485 pKa = 10.67LNATATAPLIYY496 pKa = 9.79STFLGGNGLDD506 pKa = 2.93IGYY509 pKa = 10.15SIGIDD514 pKa = 3.21AAGNAYY520 pKa = 9.54IAGFTDD526 pKa = 3.38SANYY530 pKa = 8.85PVEE533 pKa = 4.55GQIQPYY539 pKa = 9.73GGAGDD544 pKa = 4.3AMVSKK549 pKa = 10.51LNPAGTGLVYY559 pKa = 9.82STYY562 pKa = 10.73LGGIARR568 pKa = 11.84DD569 pKa = 3.44EE570 pKa = 3.97ARR572 pKa = 11.84AIVVDD577 pKa = 3.88PAGNAYY583 pKa = 8.82LTGNTEE589 pKa = 3.84SSDD592 pKa = 3.71YY593 pKa = 9.47PLSVGAIQTGLLGVRR608 pKa = 11.84DD609 pKa = 3.8VFITLFNTAGNALVYY624 pKa = 9.45STYY627 pKa = 10.98LGGTGVEE634 pKa = 4.2EE635 pKa = 4.37GLGIAVDD642 pKa = 4.24SNFNLYY648 pKa = 8.15VTGSTTSLNFPITPGAFQTSLPGLPGSGFVTKK680 pKa = 10.31IGTPIQLLGEE690 pKa = 4.34TGATGDD696 pKa = 4.18PGVTGPTGPTGDD708 pKa = 3.53TGATGEE714 pKa = 4.28TGATGEE720 pKa = 4.28TGATGEE726 pKa = 4.37TGATGDD732 pKa = 3.54TGATGEE738 pKa = 4.38TGATGDD744 pKa = 3.54TGATGEE750 pKa = 4.38TGATGDD756 pKa = 3.73TGPTGGTGATGDD768 pKa = 3.45TGATGEE774 pKa = 4.38TGATGDD780 pKa = 3.73TGPTGGTGATGDD792 pKa = 3.49TGATGGTGATGGTGATGEE810 pKa = 4.28TGATGDD816 pKa = 3.67TGATGDD822 pKa = 3.89TGATGATGEE831 pKa = 4.41TGATGDD837 pKa = 3.54TGATGEE843 pKa = 4.38TGATGDD849 pKa = 3.4TGAIGGTGATGDD861 pKa = 3.58TGATGDD867 pKa = 3.63TGATGEE873 pKa = 4.25TGATGGTGAAGEE885 pKa = 4.27TGATGEE891 pKa = 4.37TGATGDD897 pKa = 3.54TGATGEE903 pKa = 4.38TGATGDD909 pKa = 3.54TGATGEE915 pKa = 4.38TGATGDD921 pKa = 3.73TGPTGGTGATGDD933 pKa = 3.49TGATGGTGATGGTGATGEE951 pKa = 4.28TGATGDD957 pKa = 3.54TGATGEE963 pKa = 4.28TGEE966 pKa = 4.42TGATGEE972 pKa = 4.37TGATGDD978 pKa = 3.54TGATGEE984 pKa = 4.38TGATGDD990 pKa = 3.4TGAIGGTGATGDD1002 pKa = 3.58TGATGDD1008 pKa = 3.63TGATGEE1014 pKa = 4.25TGATGGTGAAGEE1026 pKa = 4.27TGATGEE1032 pKa = 4.41TGATGG1037 pKa = 3.17
Molecular weight: 101.12 kDa
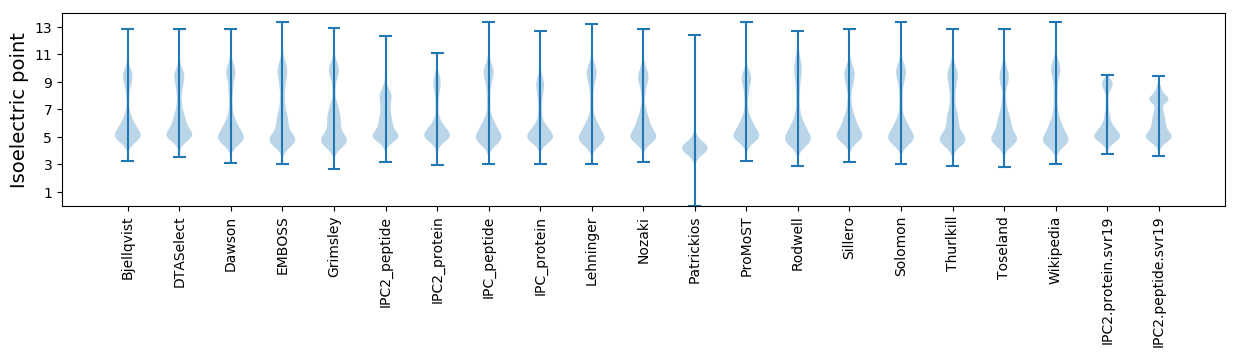
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A433YB49|A0A433YB49_9BACL CBS domain-containing protein OS=Paenibacillus anaericanus OX=170367 GN=EJP82_07905 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84PTFKK6 pKa = 10.82PNVSKK11 pKa = 10.8RR12 pKa = 11.84KK13 pKa = 8.95KK14 pKa = 8.25VHH16 pKa = 5.49GFRR19 pKa = 11.84KK20 pKa = 9.94RR21 pKa = 11.84MSTASGRR28 pKa = 11.84KK29 pKa = 8.42VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.79VLSAA44 pKa = 4.05
MM1 pKa = 7.9RR2 pKa = 11.84PTFKK6 pKa = 10.82PNVSKK11 pKa = 10.8RR12 pKa = 11.84KK13 pKa = 8.95KK14 pKa = 8.25VHH16 pKa = 5.49GFRR19 pKa = 11.84KK20 pKa = 9.94RR21 pKa = 11.84MSTASGRR28 pKa = 11.84KK29 pKa = 8.42VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.79VLSAA44 pKa = 4.05
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1728414 |
16 |
2567 |
319.2 |
35.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.011 ± 0.036 | 0.788 ± 0.011 |
5.286 ± 0.028 | 6.819 ± 0.038 |
4.248 ± 0.026 | 7.204 ± 0.041 |
2.006 ± 0.017 | 7.526 ± 0.035 |
5.769 ± 0.026 | 10.011 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.827 ± 0.017 | 4.36 ± 0.024 |
3.729 ± 0.024 | 3.78 ± 0.018 |
4.477 ± 0.028 | 6.701 ± 0.033 |
5.587 ± 0.04 | 7.003 ± 0.026 |
1.226 ± 0.014 | 3.642 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
