
Cucumber green mottle mosaic virus (strain watermelon SH) (CGMMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus; Cucumber green mottle mosaic virus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
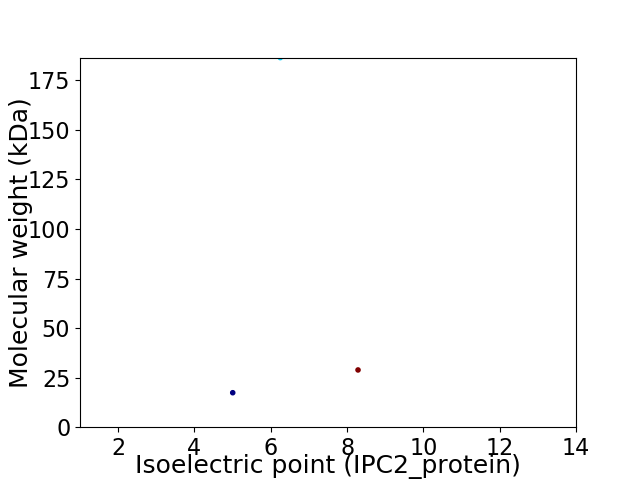
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
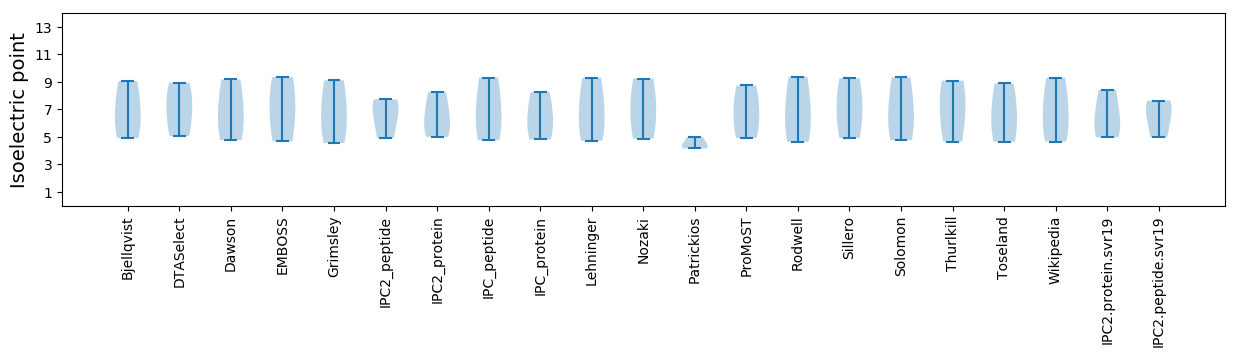
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P69514|RDRP_CGMVS Replicase large subunit OS=Cucumber green mottle mosaic virus (strain watermelon SH) OX=12236 PE=3 SV=1
MM1 pKa = 8.08AYY3 pKa = 10.67NPITPSKK10 pKa = 10.34LIAFSASYY18 pKa = 11.04VPVRR22 pKa = 11.84TLLNFLVASQGTAFQTQAGRR42 pKa = 11.84DD43 pKa = 3.58SFRR46 pKa = 11.84EE47 pKa = 4.08SLSALPSSVVDD58 pKa = 3.42INSRR62 pKa = 11.84FPDD65 pKa = 3.21AGFYY69 pKa = 10.97AFLNGPVLRR78 pKa = 11.84PIFVSLLSSTDD89 pKa = 2.69TRR91 pKa = 11.84NRR93 pKa = 11.84VIEE96 pKa = 4.2VVDD99 pKa = 3.77PSNPTTAEE107 pKa = 3.62SLNAVKK113 pKa = 9.56RR114 pKa = 11.84TDD116 pKa = 3.77DD117 pKa = 3.25ASTAARR123 pKa = 11.84AEE125 pKa = 3.78IDD127 pKa = 3.48NLIEE131 pKa = 4.58SISKK135 pKa = 10.6GFDD138 pKa = 3.05VYY140 pKa = 11.54DD141 pKa = 3.65RR142 pKa = 11.84ASFEE146 pKa = 4.19AAFSVVWSEE155 pKa = 3.89ATTSKK160 pKa = 11.24AA161 pKa = 3.27
MM1 pKa = 8.08AYY3 pKa = 10.67NPITPSKK10 pKa = 10.34LIAFSASYY18 pKa = 11.04VPVRR22 pKa = 11.84TLLNFLVASQGTAFQTQAGRR42 pKa = 11.84DD43 pKa = 3.58SFRR46 pKa = 11.84EE47 pKa = 4.08SLSALPSSVVDD58 pKa = 3.42INSRR62 pKa = 11.84FPDD65 pKa = 3.21AGFYY69 pKa = 10.97AFLNGPVLRR78 pKa = 11.84PIFVSLLSSTDD89 pKa = 2.69TRR91 pKa = 11.84NRR93 pKa = 11.84VIEE96 pKa = 4.2VVDD99 pKa = 3.77PSNPTTAEE107 pKa = 3.62SLNAVKK113 pKa = 9.56RR114 pKa = 11.84TDD116 pKa = 3.77DD117 pKa = 3.25ASTAARR123 pKa = 11.84AEE125 pKa = 3.78IDD127 pKa = 3.48NLIEE131 pKa = 4.58SISKK135 pKa = 10.6GFDD138 pKa = 3.05VYY140 pKa = 11.54DD141 pKa = 3.65RR142 pKa = 11.84ASFEE146 pKa = 4.19AAFSVVWSEE155 pKa = 3.89ATTSKK160 pKa = 11.24AA161 pKa = 3.27
Molecular weight: 17.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P69474|CAPSD_CGMVS Capsid protein OS=Cucumber green mottle mosaic virus (strain watermelon SH) OX=12236 GN=CP PE=3 SV=2
MM1 pKa = 7.34SLSKK5 pKa = 11.08VSVEE9 pKa = 4.11NSLKK13 pKa = 10.18PEE15 pKa = 4.01KK16 pKa = 9.78FVKK19 pKa = 10.4ISWVDD24 pKa = 3.32KK25 pKa = 10.65LLPNYY30 pKa = 10.05FSILKK35 pKa = 9.33YY36 pKa = 10.92LSITDD41 pKa = 3.82FSVVKK46 pKa = 9.92AQSYY50 pKa = 9.68EE51 pKa = 3.98SLVPVKK57 pKa = 10.46LLRR60 pKa = 11.84GVDD63 pKa = 3.97LTKK66 pKa = 10.58HH67 pKa = 6.35LYY69 pKa = 8.19VTLLGVVVSGVWNVPEE85 pKa = 4.23SCRR88 pKa = 11.84GGATVALVDD97 pKa = 3.72TRR99 pKa = 11.84MHH101 pKa = 6.14SVAEE105 pKa = 3.95GTICKK110 pKa = 10.07FSAPATVRR118 pKa = 11.84EE119 pKa = 4.05FSVRR123 pKa = 11.84FIPNYY128 pKa = 9.53PVVAADD134 pKa = 3.99ALRR137 pKa = 11.84DD138 pKa = 3.58PWSLFVRR145 pKa = 11.84LSNVGIKK152 pKa = 10.52DD153 pKa = 3.42GFHH156 pKa = 7.1PLTLEE161 pKa = 4.28VACLVATTNSIIKK174 pKa = 9.78KK175 pKa = 8.24GLRR178 pKa = 11.84ASVVEE183 pKa = 4.38SVVSSDD189 pKa = 3.3QSIVLDD195 pKa = 3.68SLSEE199 pKa = 4.04KK200 pKa = 10.64VEE202 pKa = 3.99PFFDD206 pKa = 3.71KK207 pKa = 11.19VPISAAVMARR217 pKa = 11.84DD218 pKa = 3.19PSYY221 pKa = 10.88RR222 pKa = 11.84SRR224 pKa = 11.84SQSVGGRR231 pKa = 11.84GKK233 pKa = 10.21RR234 pKa = 11.84HH235 pKa = 5.83SKK237 pKa = 9.55PPNRR241 pKa = 11.84RR242 pKa = 11.84LDD244 pKa = 3.66SASEE248 pKa = 4.06EE249 pKa = 4.23SSSVSFEE256 pKa = 5.18DD257 pKa = 5.04GLQSDD262 pKa = 4.21HH263 pKa = 5.96TT264 pKa = 4.23
MM1 pKa = 7.34SLSKK5 pKa = 11.08VSVEE9 pKa = 4.11NSLKK13 pKa = 10.18PEE15 pKa = 4.01KK16 pKa = 9.78FVKK19 pKa = 10.4ISWVDD24 pKa = 3.32KK25 pKa = 10.65LLPNYY30 pKa = 10.05FSILKK35 pKa = 9.33YY36 pKa = 10.92LSITDD41 pKa = 3.82FSVVKK46 pKa = 9.92AQSYY50 pKa = 9.68EE51 pKa = 3.98SLVPVKK57 pKa = 10.46LLRR60 pKa = 11.84GVDD63 pKa = 3.97LTKK66 pKa = 10.58HH67 pKa = 6.35LYY69 pKa = 8.19VTLLGVVVSGVWNVPEE85 pKa = 4.23SCRR88 pKa = 11.84GGATVALVDD97 pKa = 3.72TRR99 pKa = 11.84MHH101 pKa = 6.14SVAEE105 pKa = 3.95GTICKK110 pKa = 10.07FSAPATVRR118 pKa = 11.84EE119 pKa = 4.05FSVRR123 pKa = 11.84FIPNYY128 pKa = 9.53PVVAADD134 pKa = 3.99ALRR137 pKa = 11.84DD138 pKa = 3.58PWSLFVRR145 pKa = 11.84LSNVGIKK152 pKa = 10.52DD153 pKa = 3.42GFHH156 pKa = 7.1PLTLEE161 pKa = 4.28VACLVATTNSIIKK174 pKa = 9.78KK175 pKa = 8.24GLRR178 pKa = 11.84ASVVEE183 pKa = 4.38SVVSSDD189 pKa = 3.3QSIVLDD195 pKa = 3.68SLSEE199 pKa = 4.04KK200 pKa = 10.64VEE202 pKa = 3.99PFFDD206 pKa = 3.71KK207 pKa = 11.19VPISAAVMARR217 pKa = 11.84DD218 pKa = 3.19PSYY221 pKa = 10.88RR222 pKa = 11.84SRR224 pKa = 11.84SQSVGGRR231 pKa = 11.84GKK233 pKa = 10.21RR234 pKa = 11.84HH235 pKa = 5.83SKK237 pKa = 9.55PPNRR241 pKa = 11.84RR242 pKa = 11.84LDD244 pKa = 3.66SASEE248 pKa = 4.06EE249 pKa = 4.23SSSVSFEE256 pKa = 5.18DD257 pKa = 5.04GLQSDD262 pKa = 4.21HH263 pKa = 5.96TT264 pKa = 4.23
Molecular weight: 28.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2073 |
161 |
1648 |
691.0 |
77.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.657 ± 1.299 | 1.737 ± 0.523 |
6.561 ± 0.452 | 4.92 ± 0.135 |
5.74 ± 0.476 | 4.679 ± 0.348 |
2.315 ± 0.617 | 5.596 ± 0.667 |
6.078 ± 0.811 | 8.924 ± 0.321 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.315 ± 0.66 | 4.245 ± 0.48 |
4.197 ± 0.58 | 2.412 ± 0.366 |
5.162 ± 0.354 | 9.358 ± 2.264 |
5.499 ± 0.493 | 9.165 ± 1.426 |
0.965 ± 0.078 | 3.425 ± 0.51 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
