
Caulobacter sp. Root1455
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Caulobacter; unclassified Caulobacter
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
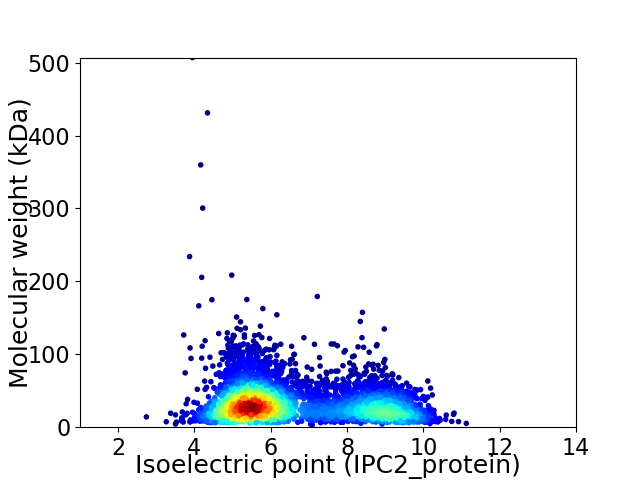
Virtual 2D-PAGE plot for 4531 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q7UF48|A0A0Q7UF48_9CAUL Methionyl-tRNA formyltransferase OS=Caulobacter sp. Root1455 OX=1736465 GN=fmt PE=3 SV=1
MM1 pKa = 6.55TTLTVGDD8 pKa = 4.44LALIGYY14 pKa = 8.84SADD17 pKa = 3.49TAGKK21 pKa = 9.52SFSFVLLQAVDD32 pKa = 3.6AGTVISFTDD41 pKa = 4.26DD42 pKa = 2.46GWLAAGGFRR51 pKa = 11.84TGEE54 pKa = 4.05GVFTYY59 pKa = 7.73TAPAGGAAAGTVITVTGLTGSLNPSTSGDD88 pKa = 3.54QIIAFQGPLSAAATPLFALDD108 pKa = 4.01FADD111 pKa = 5.49GNATYY116 pKa = 10.46AADD119 pKa = 3.72ASNSNTSAVPTGLATGSTALAFGTDD144 pKa = 2.82NGAYY148 pKa = 10.21VGTTTGTKK156 pKa = 10.12AAILSAIANSANWSLDD172 pKa = 2.98DD173 pKa = 4.87ANPVAYY179 pKa = 8.08KK180 pKa = 10.3TGFTVTDD187 pKa = 3.66GGVPSANVSISDD199 pKa = 3.57VSLTEE204 pKa = 4.25GDD206 pKa = 3.7AGTQVMTFTVTRR218 pKa = 11.84TNTTGAFTVDD228 pKa = 3.48YY229 pKa = 8.63ATADD233 pKa = 3.57GSAHH237 pKa = 6.34VGDD240 pKa = 5.69DD241 pKa = 3.36YY242 pKa = 12.19AEE244 pKa = 4.01AHH246 pKa = 5.84GTLTFAAGGPASQTISVTLNGDD268 pKa = 3.32ATPEE272 pKa = 4.08PNEE275 pKa = 4.02TFAVNLSNLVVASGAAALIDD295 pKa = 3.61AVGQATIVNDD305 pKa = 3.63DD306 pKa = 4.0FAPVAIYY313 pKa = 9.87DD314 pKa = 3.69IQGAGHH320 pKa = 6.23VSAYY324 pKa = 10.44DD325 pKa = 3.57GQTVSTQGVVTAIDD339 pKa = 3.67TTGSRR344 pKa = 11.84GFWIQDD350 pKa = 3.17ATGDD354 pKa = 4.03GDD356 pKa = 4.86DD357 pKa = 4.03ATSDD361 pKa = 3.29AVFVFTNAIPTVHH374 pKa = 6.84VGDD377 pKa = 4.6LVQVTGTVDD386 pKa = 3.31EE387 pKa = 4.71YY388 pKa = 11.69NGGVATNLSITEE400 pKa = 3.87ITAPTISVIGTGTITATVLGAGGRR424 pKa = 11.84AIPTTVIDD432 pKa = 4.84DD433 pKa = 3.83DD434 pKa = 4.71HH435 pKa = 6.53LTSFDD440 pKa = 3.74PAADD444 pKa = 3.28GVDD447 pKa = 4.1FYY449 pKa = 11.63EE450 pKa = 4.58SVEE453 pKa = 4.12GMVVTLHH460 pKa = 7.14DD461 pKa = 4.3AQSVGSTSQGQTWVVADD478 pKa = 4.29NGADD482 pKa = 3.36ASGLNSRR489 pKa = 11.84GGVTVSDD496 pKa = 3.73GDD498 pKa = 3.96NNPEE502 pKa = 4.47RR503 pKa = 11.84ILVYY507 pKa = 10.47ADD509 pKa = 3.47SGVNPGFSAGYY520 pKa = 10.15VLGDD524 pKa = 3.59HH525 pKa = 6.83LGDD528 pKa = 3.44VTGVVSYY535 pKa = 10.24FGGEE539 pKa = 4.22YY540 pKa = 10.13EE541 pKa = 4.32VLATSVQNTTSGGTLPLEE559 pKa = 4.47TTSLAGDD566 pKa = 4.09ASHH569 pKa = 7.19LAIGAYY575 pKa = 9.23NLEE578 pKa = 4.79NISPVDD584 pKa = 3.62PDD586 pKa = 3.72AKK588 pKa = 10.28FAALAADD595 pKa = 3.66IAHH598 pKa = 6.71NLGAPDD604 pKa = 3.31ILGVEE609 pKa = 5.12EE610 pKa = 4.36IQDD613 pKa = 3.39ADD615 pKa = 3.76GAGNGTNYY623 pKa = 10.63SGAATLNKK631 pKa = 9.82LVAAIEE637 pKa = 4.29AAGGPHH643 pKa = 5.95YY644 pKa = 10.65SWVEE648 pKa = 3.61IAPTANNSTGGEE660 pKa = 4.27SNGNIRR666 pKa = 11.84NAFLYY671 pKa = 10.75RR672 pKa = 11.84EE673 pKa = 4.16DD674 pKa = 3.49RR675 pKa = 11.84VDD677 pKa = 3.63YY678 pKa = 11.32VEE680 pKa = 4.77GSARR684 pKa = 11.84LIQDD688 pKa = 3.22ATGSTDD694 pKa = 2.91AFHH697 pKa = 7.63NSRR700 pKa = 11.84NPLAAEE706 pKa = 4.22FVFHH710 pKa = 7.09GEE712 pKa = 3.9TVTAIDD718 pKa = 3.18VHH720 pKa = 6.79NYY722 pKa = 9.55SRR724 pKa = 11.84GGSDD728 pKa = 3.51PLFGANQPASNNGDD742 pKa = 3.63DD743 pKa = 4.27RR744 pKa = 11.84RR745 pKa = 11.84ADD747 pKa = 3.34QSTAVHH753 pKa = 6.97DD754 pKa = 4.14YY755 pKa = 9.6VTTLLAADD763 pKa = 4.5PDD765 pKa = 3.64AHH767 pKa = 5.77ITVMGDD773 pKa = 2.96FNAYY777 pKa = 10.05YY778 pKa = 10.24YY779 pKa = 9.73EE780 pKa = 4.03QSLTLLEE787 pKa = 4.72ANGDD791 pKa = 4.07LYY793 pKa = 11.74NLARR797 pKa = 11.84TLSAEE802 pKa = 3.72EE803 pKa = 4.1RR804 pKa = 11.84YY805 pKa = 10.55SYY807 pKa = 10.68IFEE810 pKa = 4.73GNAQQIDD817 pKa = 4.06NLLVSQSLKK826 pKa = 11.02DD827 pKa = 3.34GAVFDD832 pKa = 5.26NIHH835 pKa = 6.82LNTGQAAIDD844 pKa = 3.62QPTDD848 pKa = 2.9HH849 pKa = 7.42DD850 pKa = 5.64AILALLSINTAPVATLDD867 pKa = 3.51GTFTVSEE874 pKa = 4.32DD875 pKa = 3.29AVLTVDD881 pKa = 3.6AAHH884 pKa = 6.82GVLANDD890 pKa = 3.57NDD892 pKa = 4.23ANSDD896 pKa = 3.42ALTAVLGQGPTYY908 pKa = 9.43GTLVLNADD916 pKa = 3.44GSFIYY921 pKa = 9.37TANANYY927 pKa = 10.46NGADD931 pKa = 3.16AFTYY935 pKa = 7.95TAHH938 pKa = 7.23DD939 pKa = 3.93AYY941 pKa = 10.55GGVSGVVTVQLGVEE955 pKa = 4.34AVNDD959 pKa = 4.22APTGAADD966 pKa = 3.57AASVAEE972 pKa = 4.38DD973 pKa = 3.36ASILVNVLANDD984 pKa = 3.75HH985 pKa = 7.16DD986 pKa = 4.38VDD988 pKa = 5.83LDD990 pKa = 3.87GLSIAALSGTKK1001 pKa = 9.71SALGASISIEE1011 pKa = 3.96NGQVRR1016 pKa = 11.84YY1017 pKa = 9.26AADD1020 pKa = 3.67ADD1022 pKa = 4.44GFDD1025 pKa = 3.85QLAAGHH1031 pKa = 5.83SVVDD1035 pKa = 3.82SFTYY1039 pKa = 9.72TATDD1043 pKa = 3.09GHH1045 pKa = 6.97GGFTAPITVSITVGEE1060 pKa = 4.35AGDD1063 pKa = 3.77NRR1065 pKa = 11.84TLSGNPVKK1073 pKa = 10.81ANSFTDD1079 pKa = 3.12VGEE1082 pKa = 4.12YY1083 pKa = 8.48DD1084 pKa = 3.26TTYY1087 pKa = 10.72TGGLLSDD1094 pKa = 4.6VINGGGGSDD1103 pKa = 3.84HH1104 pKa = 7.14LFGGNGKK1111 pKa = 8.45DD1112 pKa = 3.46TLNGGTGRR1120 pKa = 11.84DD1121 pKa = 3.58TLEE1124 pKa = 4.75GGTGPDD1130 pKa = 4.33LLTGGAGADD1139 pKa = 3.34DD1140 pKa = 4.24FVFGLGSGTDD1150 pKa = 3.82HH1151 pKa = 6.28ITDD1154 pKa = 4.65FNPGEE1159 pKa = 4.06DD1160 pKa = 4.4HH1161 pKa = 7.38IITPLRR1167 pKa = 11.84DD1168 pKa = 3.35QLGDD1172 pKa = 3.55FVGGLGLGKK1181 pKa = 10.28LADD1184 pKa = 3.82LVDD1187 pKa = 4.01RR1188 pKa = 11.84LLAPSFKK1195 pKa = 10.9DD1196 pKa = 3.0VDD1198 pKa = 4.15TNGDD1202 pKa = 3.11HH1203 pKa = 7.0HH1204 pKa = 7.9ADD1206 pKa = 3.18AVSINAVGMGTVVLDD1221 pKa = 3.19NWTIATLTAQGYY1233 pKa = 10.29LDD1235 pKa = 4.26PQHH1238 pKa = 6.76HH1239 pKa = 6.6VVGDD1243 pKa = 3.58WLLL1246 pKa = 3.58
MM1 pKa = 6.55TTLTVGDD8 pKa = 4.44LALIGYY14 pKa = 8.84SADD17 pKa = 3.49TAGKK21 pKa = 9.52SFSFVLLQAVDD32 pKa = 3.6AGTVISFTDD41 pKa = 4.26DD42 pKa = 2.46GWLAAGGFRR51 pKa = 11.84TGEE54 pKa = 4.05GVFTYY59 pKa = 7.73TAPAGGAAAGTVITVTGLTGSLNPSTSGDD88 pKa = 3.54QIIAFQGPLSAAATPLFALDD108 pKa = 4.01FADD111 pKa = 5.49GNATYY116 pKa = 10.46AADD119 pKa = 3.72ASNSNTSAVPTGLATGSTALAFGTDD144 pKa = 2.82NGAYY148 pKa = 10.21VGTTTGTKK156 pKa = 10.12AAILSAIANSANWSLDD172 pKa = 2.98DD173 pKa = 4.87ANPVAYY179 pKa = 8.08KK180 pKa = 10.3TGFTVTDD187 pKa = 3.66GGVPSANVSISDD199 pKa = 3.57VSLTEE204 pKa = 4.25GDD206 pKa = 3.7AGTQVMTFTVTRR218 pKa = 11.84TNTTGAFTVDD228 pKa = 3.48YY229 pKa = 8.63ATADD233 pKa = 3.57GSAHH237 pKa = 6.34VGDD240 pKa = 5.69DD241 pKa = 3.36YY242 pKa = 12.19AEE244 pKa = 4.01AHH246 pKa = 5.84GTLTFAAGGPASQTISVTLNGDD268 pKa = 3.32ATPEE272 pKa = 4.08PNEE275 pKa = 4.02TFAVNLSNLVVASGAAALIDD295 pKa = 3.61AVGQATIVNDD305 pKa = 3.63DD306 pKa = 4.0FAPVAIYY313 pKa = 9.87DD314 pKa = 3.69IQGAGHH320 pKa = 6.23VSAYY324 pKa = 10.44DD325 pKa = 3.57GQTVSTQGVVTAIDD339 pKa = 3.67TTGSRR344 pKa = 11.84GFWIQDD350 pKa = 3.17ATGDD354 pKa = 4.03GDD356 pKa = 4.86DD357 pKa = 4.03ATSDD361 pKa = 3.29AVFVFTNAIPTVHH374 pKa = 6.84VGDD377 pKa = 4.6LVQVTGTVDD386 pKa = 3.31EE387 pKa = 4.71YY388 pKa = 11.69NGGVATNLSITEE400 pKa = 3.87ITAPTISVIGTGTITATVLGAGGRR424 pKa = 11.84AIPTTVIDD432 pKa = 4.84DD433 pKa = 3.83DD434 pKa = 4.71HH435 pKa = 6.53LTSFDD440 pKa = 3.74PAADD444 pKa = 3.28GVDD447 pKa = 4.1FYY449 pKa = 11.63EE450 pKa = 4.58SVEE453 pKa = 4.12GMVVTLHH460 pKa = 7.14DD461 pKa = 4.3AQSVGSTSQGQTWVVADD478 pKa = 4.29NGADD482 pKa = 3.36ASGLNSRR489 pKa = 11.84GGVTVSDD496 pKa = 3.73GDD498 pKa = 3.96NNPEE502 pKa = 4.47RR503 pKa = 11.84ILVYY507 pKa = 10.47ADD509 pKa = 3.47SGVNPGFSAGYY520 pKa = 10.15VLGDD524 pKa = 3.59HH525 pKa = 6.83LGDD528 pKa = 3.44VTGVVSYY535 pKa = 10.24FGGEE539 pKa = 4.22YY540 pKa = 10.13EE541 pKa = 4.32VLATSVQNTTSGGTLPLEE559 pKa = 4.47TTSLAGDD566 pKa = 4.09ASHH569 pKa = 7.19LAIGAYY575 pKa = 9.23NLEE578 pKa = 4.79NISPVDD584 pKa = 3.62PDD586 pKa = 3.72AKK588 pKa = 10.28FAALAADD595 pKa = 3.66IAHH598 pKa = 6.71NLGAPDD604 pKa = 3.31ILGVEE609 pKa = 5.12EE610 pKa = 4.36IQDD613 pKa = 3.39ADD615 pKa = 3.76GAGNGTNYY623 pKa = 10.63SGAATLNKK631 pKa = 9.82LVAAIEE637 pKa = 4.29AAGGPHH643 pKa = 5.95YY644 pKa = 10.65SWVEE648 pKa = 3.61IAPTANNSTGGEE660 pKa = 4.27SNGNIRR666 pKa = 11.84NAFLYY671 pKa = 10.75RR672 pKa = 11.84EE673 pKa = 4.16DD674 pKa = 3.49RR675 pKa = 11.84VDD677 pKa = 3.63YY678 pKa = 11.32VEE680 pKa = 4.77GSARR684 pKa = 11.84LIQDD688 pKa = 3.22ATGSTDD694 pKa = 2.91AFHH697 pKa = 7.63NSRR700 pKa = 11.84NPLAAEE706 pKa = 4.22FVFHH710 pKa = 7.09GEE712 pKa = 3.9TVTAIDD718 pKa = 3.18VHH720 pKa = 6.79NYY722 pKa = 9.55SRR724 pKa = 11.84GGSDD728 pKa = 3.51PLFGANQPASNNGDD742 pKa = 3.63DD743 pKa = 4.27RR744 pKa = 11.84RR745 pKa = 11.84ADD747 pKa = 3.34QSTAVHH753 pKa = 6.97DD754 pKa = 4.14YY755 pKa = 9.6VTTLLAADD763 pKa = 4.5PDD765 pKa = 3.64AHH767 pKa = 5.77ITVMGDD773 pKa = 2.96FNAYY777 pKa = 10.05YY778 pKa = 10.24YY779 pKa = 9.73EE780 pKa = 4.03QSLTLLEE787 pKa = 4.72ANGDD791 pKa = 4.07LYY793 pKa = 11.74NLARR797 pKa = 11.84TLSAEE802 pKa = 3.72EE803 pKa = 4.1RR804 pKa = 11.84YY805 pKa = 10.55SYY807 pKa = 10.68IFEE810 pKa = 4.73GNAQQIDD817 pKa = 4.06NLLVSQSLKK826 pKa = 11.02DD827 pKa = 3.34GAVFDD832 pKa = 5.26NIHH835 pKa = 6.82LNTGQAAIDD844 pKa = 3.62QPTDD848 pKa = 2.9HH849 pKa = 7.42DD850 pKa = 5.64AILALLSINTAPVATLDD867 pKa = 3.51GTFTVSEE874 pKa = 4.32DD875 pKa = 3.29AVLTVDD881 pKa = 3.6AAHH884 pKa = 6.82GVLANDD890 pKa = 3.57NDD892 pKa = 4.23ANSDD896 pKa = 3.42ALTAVLGQGPTYY908 pKa = 9.43GTLVLNADD916 pKa = 3.44GSFIYY921 pKa = 9.37TANANYY927 pKa = 10.46NGADD931 pKa = 3.16AFTYY935 pKa = 7.95TAHH938 pKa = 7.23DD939 pKa = 3.93AYY941 pKa = 10.55GGVSGVVTVQLGVEE955 pKa = 4.34AVNDD959 pKa = 4.22APTGAADD966 pKa = 3.57AASVAEE972 pKa = 4.38DD973 pKa = 3.36ASILVNVLANDD984 pKa = 3.75HH985 pKa = 7.16DD986 pKa = 4.38VDD988 pKa = 5.83LDD990 pKa = 3.87GLSIAALSGTKK1001 pKa = 9.71SALGASISIEE1011 pKa = 3.96NGQVRR1016 pKa = 11.84YY1017 pKa = 9.26AADD1020 pKa = 3.67ADD1022 pKa = 4.44GFDD1025 pKa = 3.85QLAAGHH1031 pKa = 5.83SVVDD1035 pKa = 3.82SFTYY1039 pKa = 9.72TATDD1043 pKa = 3.09GHH1045 pKa = 6.97GGFTAPITVSITVGEE1060 pKa = 4.35AGDD1063 pKa = 3.77NRR1065 pKa = 11.84TLSGNPVKK1073 pKa = 10.81ANSFTDD1079 pKa = 3.12VGEE1082 pKa = 4.12YY1083 pKa = 8.48DD1084 pKa = 3.26TTYY1087 pKa = 10.72TGGLLSDD1094 pKa = 4.6VINGGGGSDD1103 pKa = 3.84HH1104 pKa = 7.14LFGGNGKK1111 pKa = 8.45DD1112 pKa = 3.46TLNGGTGRR1120 pKa = 11.84DD1121 pKa = 3.58TLEE1124 pKa = 4.75GGTGPDD1130 pKa = 4.33LLTGGAGADD1139 pKa = 3.34DD1140 pKa = 4.24FVFGLGSGTDD1150 pKa = 3.82HH1151 pKa = 6.28ITDD1154 pKa = 4.65FNPGEE1159 pKa = 4.06DD1160 pKa = 4.4HH1161 pKa = 7.38IITPLRR1167 pKa = 11.84DD1168 pKa = 3.35QLGDD1172 pKa = 3.55FVGGLGLGKK1181 pKa = 10.28LADD1184 pKa = 3.82LVDD1187 pKa = 4.01RR1188 pKa = 11.84LLAPSFKK1195 pKa = 10.9DD1196 pKa = 3.0VDD1198 pKa = 4.15TNGDD1202 pKa = 3.11HH1203 pKa = 7.0HH1204 pKa = 7.9ADD1206 pKa = 3.18AVSINAVGMGTVVLDD1221 pKa = 3.19NWTIATLTAQGYY1233 pKa = 10.29LDD1235 pKa = 4.26PQHH1238 pKa = 6.76HH1239 pKa = 6.6VVGDD1243 pKa = 3.58WLLL1246 pKa = 3.58
Molecular weight: 126.48 kDa
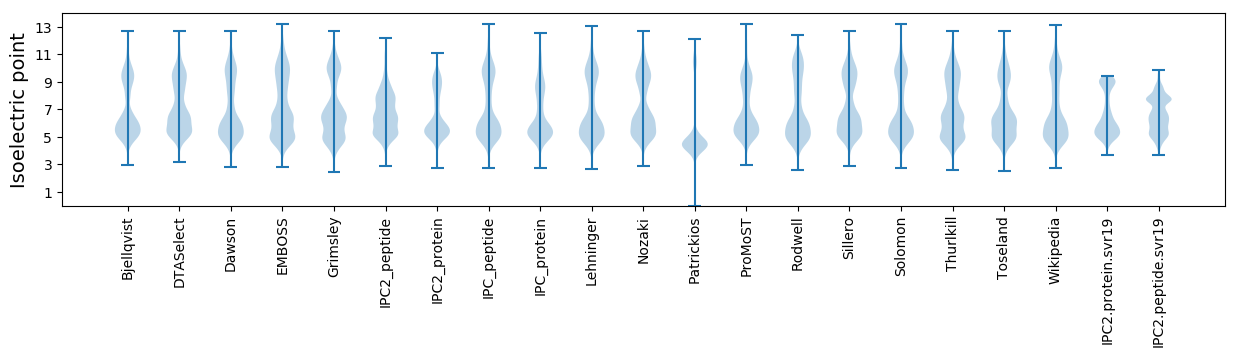
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q7TIU5|A0A0Q7TIU5_9CAUL Chemotaxis protein OS=Caulobacter sp. Root1455 OX=1736465 GN=ASD21_22400 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.58LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 5.26GYY18 pKa = 7.96RR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.84NGQKK29 pKa = 9.53IVARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.58LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 5.26GYY18 pKa = 7.96RR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.84NGQKK29 pKa = 9.53IVARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1466033 |
29 |
4887 |
323.6 |
34.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.936 ± 0.057 | 0.735 ± 0.012 |
5.98 ± 0.029 | 5.148 ± 0.038 |
3.578 ± 0.023 | 9.131 ± 0.07 |
1.817 ± 0.017 | 4.273 ± 0.021 |
3.36 ± 0.029 | 10.009 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.16 ± 0.019 | 2.393 ± 0.028 |
5.618 ± 0.035 | 3.094 ± 0.023 |
6.936 ± 0.046 | 5.046 ± 0.034 |
5.409 ± 0.04 | 7.683 ± 0.029 |
1.471 ± 0.016 | 2.222 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
