
Pneumocystis jirovecii (strain RU7) (Human pneumocystis pneumonia agent)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; Taphrinomycotina; Pneumocystidomycetes; Pneumocystidales; Pneumocystidaceae; Pneumocystis; Pneumocystis jirovecii
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
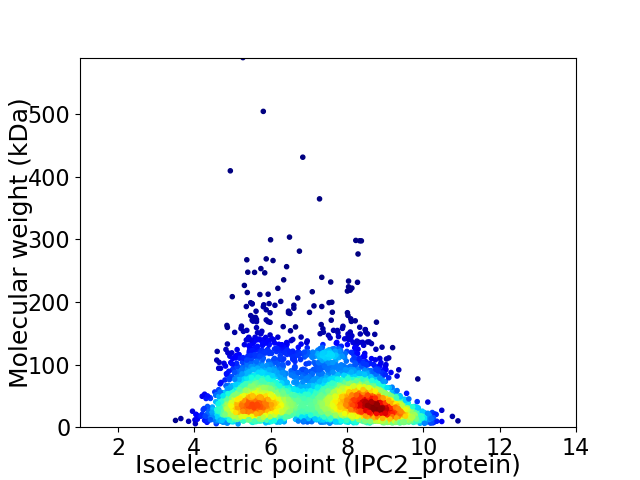
Virtual 2D-PAGE plot for 3746 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W4ZV09|A0A0W4ZV09_PNEJ7 4F5 domain-containing protein OS=Pneumocystis jirovecii (strain RU7) OX=1408657 GN=T551_00896 PE=4 SV=1
MM1 pKa = 7.43SFYY4 pKa = 11.25DD5 pKa = 4.0EE6 pKa = 5.34IEE8 pKa = 4.56IEE10 pKa = 5.03DD11 pKa = 3.75FTFDD15 pKa = 3.35TEE17 pKa = 4.2LQIYY21 pKa = 8.98HH22 pKa = 5.9YY23 pKa = 9.19PCPCGDD29 pKa = 3.35RR30 pKa = 11.84FEE32 pKa = 5.82ISLNDD37 pKa = 3.47LRR39 pKa = 11.84KK40 pKa = 9.65GDD42 pKa = 4.37EE43 pKa = 3.73IAYY46 pKa = 9.97CPSCSLMVRR55 pKa = 11.84VIYY58 pKa = 10.58DD59 pKa = 3.06QDD61 pKa = 3.41KK62 pKa = 10.55FLEE65 pKa = 4.34NDD67 pKa = 4.14DD68 pKa = 4.82EE69 pKa = 5.89ISLQSAPFIVVV80 pKa = 3.34
MM1 pKa = 7.43SFYY4 pKa = 11.25DD5 pKa = 4.0EE6 pKa = 5.34IEE8 pKa = 4.56IEE10 pKa = 5.03DD11 pKa = 3.75FTFDD15 pKa = 3.35TEE17 pKa = 4.2LQIYY21 pKa = 8.98HH22 pKa = 5.9YY23 pKa = 9.19PCPCGDD29 pKa = 3.35RR30 pKa = 11.84FEE32 pKa = 5.82ISLNDD37 pKa = 3.47LRR39 pKa = 11.84KK40 pKa = 9.65GDD42 pKa = 4.37EE43 pKa = 3.73IAYY46 pKa = 9.97CPSCSLMVRR55 pKa = 11.84VIYY58 pKa = 10.58DD59 pKa = 3.06QDD61 pKa = 3.41KK62 pKa = 10.55FLEE65 pKa = 4.34NDD67 pKa = 4.14DD68 pKa = 4.82EE69 pKa = 5.89ISLQSAPFIVVV80 pKa = 3.34
Molecular weight: 9.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W4ZD85|A0A0W4ZD85_PNEJ7 Ubiquitin-60S ribosomal protein L40 OS=Pneumocystis jirovecii (strain RU7) OX=1408657 GN=T551_03569 PE=4 SV=1
MM1 pKa = 7.44SLHH4 pKa = 6.81ALHH7 pKa = 7.0ALHH10 pKa = 7.17ASHH13 pKa = 6.83APKK16 pKa = 10.26RR17 pKa = 11.84ALFLRR22 pKa = 11.84GPSSSPALHH31 pKa = 7.15RR32 pKa = 11.84ILADD36 pKa = 4.16LSALAPDD43 pKa = 4.15SVSLSRR49 pKa = 11.84PNALYY54 pKa = 10.03PFDD57 pKa = 5.79DD58 pKa = 4.35PASLEE63 pKa = 4.03FLAHH67 pKa = 6.57KK68 pKa = 10.24NHH70 pKa = 6.67AALVVVASSSRR81 pKa = 11.84KK82 pKa = 9.82RR83 pKa = 11.84GDD85 pKa = 3.22VLIFARR91 pKa = 11.84LFDD94 pKa = 4.17GALLDD99 pKa = 3.8MLEE102 pKa = 4.58LCLSDD107 pKa = 3.73AHH109 pKa = 6.41PMHH112 pKa = 6.95LFSAKK117 pKa = 9.97SPCGLRR123 pKa = 11.84PLLLFAGAPFDD134 pKa = 3.63VSPPHH139 pKa = 6.57RR140 pKa = 11.84LCSSLFLDD148 pKa = 5.02LFGGPLPRR156 pKa = 11.84IHH158 pKa = 7.67ADD160 pKa = 3.35GLQHH164 pKa = 7.0AVCLTATDD172 pKa = 3.47SHH174 pKa = 6.54AHH176 pKa = 5.23LRR178 pKa = 11.84VYY180 pKa = 7.4TLCRR184 pKa = 11.84GATSVSLCEE193 pKa = 4.02AGPRR197 pKa = 11.84LDD199 pKa = 3.59FTLGRR204 pKa = 11.84VRR206 pKa = 11.84EE207 pKa = 4.01ASAAQLHH214 pKa = 6.54AATAPAAVPRR224 pKa = 11.84PPRR227 pKa = 11.84NTSLSSRR234 pKa = 11.84GDD236 pKa = 3.07RR237 pKa = 11.84VGRR240 pKa = 11.84VHH242 pKa = 6.63VAQRR246 pKa = 11.84LSLQTRR252 pKa = 11.84KK253 pKa = 9.28FRR255 pKa = 11.84ALRR258 pKa = 11.84ARR260 pKa = 11.84RR261 pKa = 11.84TGGG264 pKa = 3.07
MM1 pKa = 7.44SLHH4 pKa = 6.81ALHH7 pKa = 7.0ALHH10 pKa = 7.17ASHH13 pKa = 6.83APKK16 pKa = 10.26RR17 pKa = 11.84ALFLRR22 pKa = 11.84GPSSSPALHH31 pKa = 7.15RR32 pKa = 11.84ILADD36 pKa = 4.16LSALAPDD43 pKa = 4.15SVSLSRR49 pKa = 11.84PNALYY54 pKa = 10.03PFDD57 pKa = 5.79DD58 pKa = 4.35PASLEE63 pKa = 4.03FLAHH67 pKa = 6.57KK68 pKa = 10.24NHH70 pKa = 6.67AALVVVASSSRR81 pKa = 11.84KK82 pKa = 9.82RR83 pKa = 11.84GDD85 pKa = 3.22VLIFARR91 pKa = 11.84LFDD94 pKa = 4.17GALLDD99 pKa = 3.8MLEE102 pKa = 4.58LCLSDD107 pKa = 3.73AHH109 pKa = 6.41PMHH112 pKa = 6.95LFSAKK117 pKa = 9.97SPCGLRR123 pKa = 11.84PLLLFAGAPFDD134 pKa = 3.63VSPPHH139 pKa = 6.57RR140 pKa = 11.84LCSSLFLDD148 pKa = 5.02LFGGPLPRR156 pKa = 11.84IHH158 pKa = 7.67ADD160 pKa = 3.35GLQHH164 pKa = 7.0AVCLTATDD172 pKa = 3.47SHH174 pKa = 6.54AHH176 pKa = 5.23LRR178 pKa = 11.84VYY180 pKa = 7.4TLCRR184 pKa = 11.84GATSVSLCEE193 pKa = 4.02AGPRR197 pKa = 11.84LDD199 pKa = 3.59FTLGRR204 pKa = 11.84VRR206 pKa = 11.84EE207 pKa = 4.01ASAAQLHH214 pKa = 6.54AATAPAAVPRR224 pKa = 11.84PPRR227 pKa = 11.84NTSLSSRR234 pKa = 11.84GDD236 pKa = 3.07RR237 pKa = 11.84VGRR240 pKa = 11.84VHH242 pKa = 6.63VAQRR246 pKa = 11.84LSLQTRR252 pKa = 11.84KK253 pKa = 9.28FRR255 pKa = 11.84ALRR258 pKa = 11.84ARR260 pKa = 11.84RR261 pKa = 11.84TGGG264 pKa = 3.07
Molecular weight: 28.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1771070 |
46 |
5096 |
472.8 |
54.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.512 ± 0.031 | 1.885 ± 0.029 |
5.381 ± 0.022 | 6.987 ± 0.041 |
4.706 ± 0.03 | 4.306 ± 0.036 |
2.228 ± 0.015 | 8.059 ± 0.051 |
8.575 ± 0.055 | 9.864 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.054 ± 0.013 | 6.179 ± 0.034 |
3.656 ± 0.03 | 3.701 ± 0.025 |
4.395 ± 0.025 | 8.538 ± 0.04 |
5.321 ± 0.031 | 4.957 ± 0.026 |
1.062 ± 0.011 | 3.634 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
