
Chickpea yellow dwarf virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
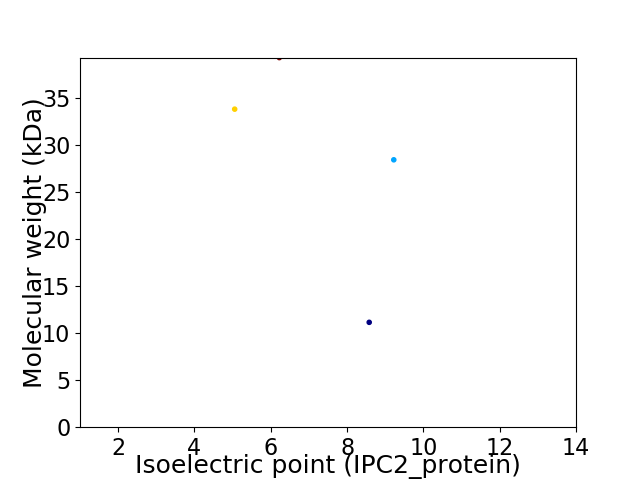
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0UF55|A0A0A0UF55_9GEMI Replication-associated protein OS=Chickpea yellow dwarf virus OX=1568974 PE=3 SV=1
MM1 pKa = 7.96PRR3 pKa = 11.84RR4 pKa = 11.84TNNNSFRR11 pKa = 11.84LQTKK15 pKa = 9.03YY16 pKa = 11.1VFLTYY21 pKa = 9.65PHH23 pKa = 6.74SSSTAEE29 pKa = 3.99NLRR32 pKa = 11.84DD33 pKa = 3.88FLWSKK38 pKa = 9.02LTSLAIFFIAIATEE52 pKa = 4.05LHH54 pKa = 6.12QDD56 pKa = 3.96GSPHH60 pKa = 5.94LHH62 pKa = 6.98CLIQLDD68 pKa = 4.15KK69 pKa = 11.07RR70 pKa = 11.84CDD72 pKa = 3.2IRR74 pKa = 11.84DD75 pKa = 3.3PSFFDD80 pKa = 4.7FEE82 pKa = 4.9GNHH85 pKa = 6.78PNIQPAKK92 pKa = 9.43NSKK95 pKa = 9.78QVLEE99 pKa = 4.68YY100 pKa = 10.19ISKK103 pKa = 10.52DD104 pKa = 3.12GNIITRR110 pKa = 11.84GDD112 pKa = 3.55FRR114 pKa = 11.84NHH116 pKa = 5.79RR117 pKa = 11.84VSPTKK122 pKa = 10.37SDD124 pKa = 3.41EE125 pKa = 3.8RR126 pKa = 11.84WRR128 pKa = 11.84TIIQTATSKK137 pKa = 11.14EE138 pKa = 4.22EE139 pKa = 3.73YY140 pKa = 10.77LEE142 pKa = 4.31MIKK145 pKa = 10.12QEE147 pKa = 4.85FPHH150 pKa = 6.17EE151 pKa = 4.21WATKK155 pKa = 8.46LHH157 pKa = 5.33WLEE160 pKa = 4.13YY161 pKa = 9.8SASKK165 pKa = 10.35LFPDD169 pKa = 4.83IEE171 pKa = 4.43PPYY174 pKa = 10.38QSPFPDD180 pKa = 3.39EE181 pKa = 4.76FLHH184 pKa = 5.57CHH186 pKa = 5.47EE187 pKa = 6.07EE188 pKa = 3.66ITEE191 pKa = 3.97WLNRR195 pKa = 11.84DD196 pKa = 3.89LYY198 pKa = 10.6VVSVDD203 pKa = 4.1SYY205 pKa = 11.68QLLHH209 pKa = 6.46PHH211 pKa = 7.01LSLEE215 pKa = 4.38TATEE219 pKa = 3.91DD220 pKa = 5.1LIWMDD225 pKa = 4.75DD226 pKa = 3.6YY227 pKa = 11.67TRR229 pKa = 11.84NPNNSNSGEE238 pKa = 4.14DD239 pKa = 3.66PSTSADD245 pKa = 3.2QVVPEE250 pKa = 4.58RR251 pKa = 11.84PVGPEE256 pKa = 3.28VWEE259 pKa = 4.31DD260 pKa = 3.53TTTSTEE266 pKa = 4.12GSISLHH272 pKa = 4.7TTSTQHH278 pKa = 5.2TTSSTTSPSSSAPTGSSS295 pKa = 2.86
MM1 pKa = 7.96PRR3 pKa = 11.84RR4 pKa = 11.84TNNNSFRR11 pKa = 11.84LQTKK15 pKa = 9.03YY16 pKa = 11.1VFLTYY21 pKa = 9.65PHH23 pKa = 6.74SSSTAEE29 pKa = 3.99NLRR32 pKa = 11.84DD33 pKa = 3.88FLWSKK38 pKa = 9.02LTSLAIFFIAIATEE52 pKa = 4.05LHH54 pKa = 6.12QDD56 pKa = 3.96GSPHH60 pKa = 5.94LHH62 pKa = 6.98CLIQLDD68 pKa = 4.15KK69 pKa = 11.07RR70 pKa = 11.84CDD72 pKa = 3.2IRR74 pKa = 11.84DD75 pKa = 3.3PSFFDD80 pKa = 4.7FEE82 pKa = 4.9GNHH85 pKa = 6.78PNIQPAKK92 pKa = 9.43NSKK95 pKa = 9.78QVLEE99 pKa = 4.68YY100 pKa = 10.19ISKK103 pKa = 10.52DD104 pKa = 3.12GNIITRR110 pKa = 11.84GDD112 pKa = 3.55FRR114 pKa = 11.84NHH116 pKa = 5.79RR117 pKa = 11.84VSPTKK122 pKa = 10.37SDD124 pKa = 3.41EE125 pKa = 3.8RR126 pKa = 11.84WRR128 pKa = 11.84TIIQTATSKK137 pKa = 11.14EE138 pKa = 4.22EE139 pKa = 3.73YY140 pKa = 10.77LEE142 pKa = 4.31MIKK145 pKa = 10.12QEE147 pKa = 4.85FPHH150 pKa = 6.17EE151 pKa = 4.21WATKK155 pKa = 8.46LHH157 pKa = 5.33WLEE160 pKa = 4.13YY161 pKa = 9.8SASKK165 pKa = 10.35LFPDD169 pKa = 4.83IEE171 pKa = 4.43PPYY174 pKa = 10.38QSPFPDD180 pKa = 3.39EE181 pKa = 4.76FLHH184 pKa = 5.57CHH186 pKa = 5.47EE187 pKa = 6.07EE188 pKa = 3.66ITEE191 pKa = 3.97WLNRR195 pKa = 11.84DD196 pKa = 3.89LYY198 pKa = 10.6VVSVDD203 pKa = 4.1SYY205 pKa = 11.68QLLHH209 pKa = 6.46PHH211 pKa = 7.01LSLEE215 pKa = 4.38TATEE219 pKa = 3.91DD220 pKa = 5.1LIWMDD225 pKa = 4.75DD226 pKa = 3.6YY227 pKa = 11.67TRR229 pKa = 11.84NPNNSNSGEE238 pKa = 4.14DD239 pKa = 3.66PSTSADD245 pKa = 3.2QVVPEE250 pKa = 4.58RR251 pKa = 11.84PVGPEE256 pKa = 3.28VWEE259 pKa = 4.31DD260 pKa = 3.53TTTSTEE266 pKa = 4.12GSISLHH272 pKa = 4.7TTSTQHH278 pKa = 5.2TTSSTTSPSSSAPTGSSS295 pKa = 2.86
Molecular weight: 33.86 kDa
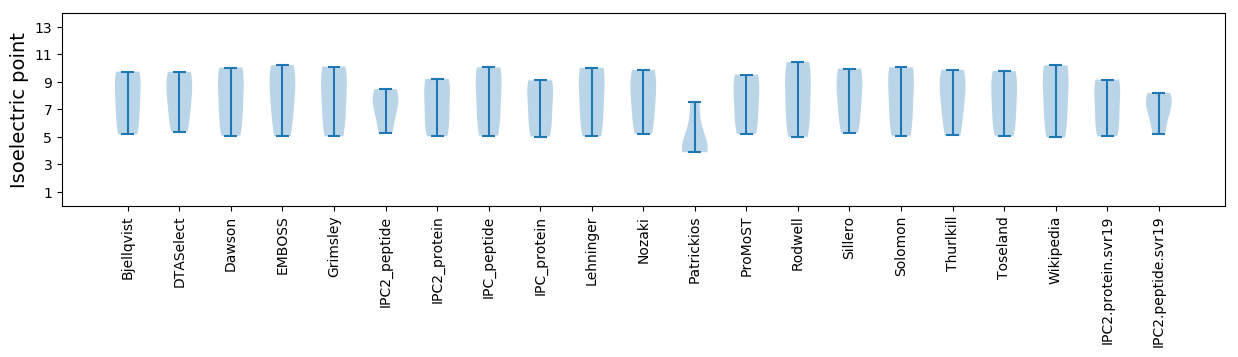
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0U8Q7|A0A0A0U8Q7_9GEMI Replication-associated protein OS=Chickpea yellow dwarf virus OX=1568974 PE=3 SV=1
MM1 pKa = 7.93PGPFSGKK8 pKa = 8.25TYY10 pKa = 10.28SRR12 pKa = 11.84KK13 pKa = 8.86KK14 pKa = 10.22GKK16 pKa = 8.59YY17 pKa = 7.81AKK19 pKa = 9.89AYY21 pKa = 8.61KK22 pKa = 10.5ALGVKK27 pKa = 7.95NQKK30 pKa = 8.75EE31 pKa = 4.13LEE33 pKa = 4.19EE34 pKa = 4.38LVNAPACPVTPRR46 pKa = 11.84PALQVAEE53 pKa = 4.51YY54 pKa = 10.17FWTTDD59 pKa = 2.93KK60 pKa = 11.59NGMIFRR66 pKa = 11.84SGGGTAHH73 pKa = 6.29FTMYY77 pKa = 10.3PQGSNEE83 pKa = 3.93NCRR86 pKa = 11.84HH87 pKa = 5.44SNQTNTYY94 pKa = 10.01KK95 pKa = 10.12MAIKK99 pKa = 10.04CWVALDD105 pKa = 3.32PTFYY109 pKa = 11.0KK110 pKa = 10.48KK111 pKa = 10.05VACVPVHH118 pKa = 6.13FWLVYY123 pKa = 10.81DD124 pKa = 4.57KK125 pKa = 11.45DD126 pKa = 4.0PGNTLPGCSTIFDD139 pKa = 4.39TLYY142 pKa = 10.62QDD144 pKa = 5.17YY145 pKa = 9.93PGTWTVSRR153 pKa = 11.84NVSRR157 pKa = 11.84RR158 pKa = 11.84FVVKK162 pKa = 9.84KK163 pKa = 8.83HH164 pKa = 4.1WHH166 pKa = 6.64IILASNGTNPTQDD179 pKa = 3.16QDD181 pKa = 3.62PAKK184 pKa = 9.85YY185 pKa = 9.91AGPGPVFQWKK195 pKa = 9.76HH196 pKa = 3.83MNKK199 pKa = 8.99FFKK202 pKa = 10.59RR203 pKa = 11.84LGVRR207 pKa = 11.84TDD209 pKa = 3.17WKK211 pKa = 10.78NSATGEE217 pKa = 4.17VADD220 pKa = 4.34IKK222 pKa = 11.13SGALYY227 pKa = 9.62LVCAPSGGAVVRR239 pKa = 11.84VGGRR243 pKa = 11.84FRR245 pKa = 11.84MYY247 pKa = 10.18FKK249 pKa = 10.96SVGNQQ254 pKa = 2.8
MM1 pKa = 7.93PGPFSGKK8 pKa = 8.25TYY10 pKa = 10.28SRR12 pKa = 11.84KK13 pKa = 8.86KK14 pKa = 10.22GKK16 pKa = 8.59YY17 pKa = 7.81AKK19 pKa = 9.89AYY21 pKa = 8.61KK22 pKa = 10.5ALGVKK27 pKa = 7.95NQKK30 pKa = 8.75EE31 pKa = 4.13LEE33 pKa = 4.19EE34 pKa = 4.38LVNAPACPVTPRR46 pKa = 11.84PALQVAEE53 pKa = 4.51YY54 pKa = 10.17FWTTDD59 pKa = 2.93KK60 pKa = 11.59NGMIFRR66 pKa = 11.84SGGGTAHH73 pKa = 6.29FTMYY77 pKa = 10.3PQGSNEE83 pKa = 3.93NCRR86 pKa = 11.84HH87 pKa = 5.44SNQTNTYY94 pKa = 10.01KK95 pKa = 10.12MAIKK99 pKa = 10.04CWVALDD105 pKa = 3.32PTFYY109 pKa = 11.0KK110 pKa = 10.48KK111 pKa = 10.05VACVPVHH118 pKa = 6.13FWLVYY123 pKa = 10.81DD124 pKa = 4.57KK125 pKa = 11.45DD126 pKa = 4.0PGNTLPGCSTIFDD139 pKa = 4.39TLYY142 pKa = 10.62QDD144 pKa = 5.17YY145 pKa = 9.93PGTWTVSRR153 pKa = 11.84NVSRR157 pKa = 11.84RR158 pKa = 11.84FVVKK162 pKa = 9.84KK163 pKa = 8.83HH164 pKa = 4.1WHH166 pKa = 6.64IILASNGTNPTQDD179 pKa = 3.16QDD181 pKa = 3.62PAKK184 pKa = 9.85YY185 pKa = 9.91AGPGPVFQWKK195 pKa = 9.76HH196 pKa = 3.83MNKK199 pKa = 8.99FFKK202 pKa = 10.59RR203 pKa = 11.84LGVRR207 pKa = 11.84TDD209 pKa = 3.17WKK211 pKa = 10.78NSATGEE217 pKa = 4.17VADD220 pKa = 4.34IKK222 pKa = 11.13SGALYY227 pKa = 9.62LVCAPSGGAVVRR239 pKa = 11.84VGGRR243 pKa = 11.84FRR245 pKa = 11.84MYY247 pKa = 10.18FKK249 pKa = 10.96SVGNQQ254 pKa = 2.8
Molecular weight: 28.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
986 |
101 |
336 |
246.5 |
28.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.97 ± 0.849 | 1.724 ± 0.297 |
5.172 ± 0.654 | 5.781 ± 1.129 |
5.274 ± 0.353 | 5.375 ± 1.321 |
3.448 ± 0.553 | 5.172 ± 1.026 |
6.187 ± 1.015 | 7.099 ± 0.728 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.42 ± 0.289 | 5.274 ± 0.211 |
6.389 ± 0.345 | 3.753 ± 0.074 |
5.071 ± 0.358 | 8.418 ± 1.299 |
7.708 ± 0.878 | 5.274 ± 1.286 |
2.231 ± 0.29 | 4.26 ± 0.446 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
