
Metschnikowia aff. pulcherrima
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Metschnikowiaceae; Metschnikowia
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5795 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
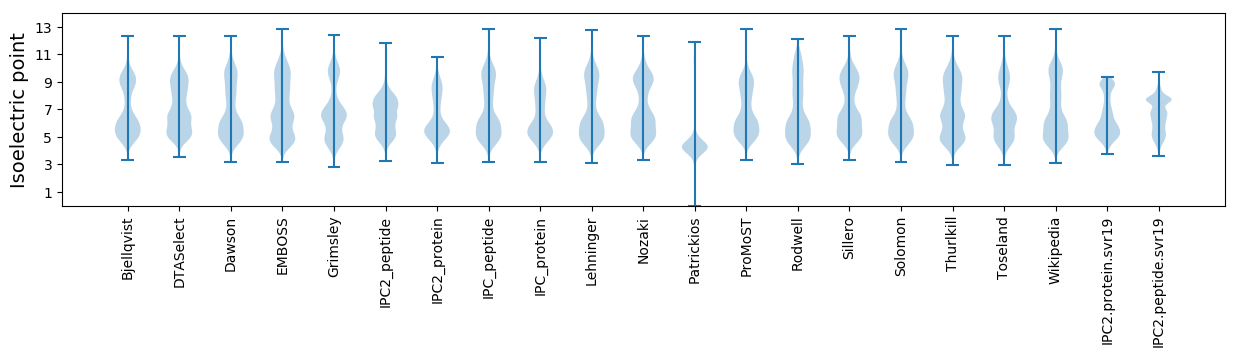
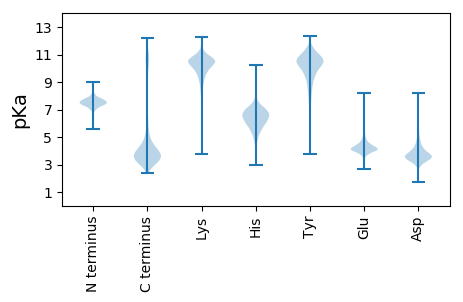
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P6XPW9|A0A4P6XPW9_9ASCO Post-GPI attachment to proteins factor 3 OS=Metschnikowia aff. pulcherrima OX=2163413 GN=MPUL0D06180 PE=3 SV=1
MM1 pKa = 6.97KK2 pKa = 9.96TGIGALATAFMCLFGVISAVTITQNTVEE30 pKa = 4.34VSPISLSLGEE40 pKa = 5.13LYY42 pKa = 10.46INPGVYY48 pKa = 10.4YY49 pKa = 10.51SIVNNFQTLLARR61 pKa = 11.84SVYY64 pKa = 10.62NKK66 pKa = 10.43GGLFITAADD75 pKa = 4.07SYY77 pKa = 8.8TASVTMTGDD86 pKa = 2.85TFMNSGIVSFNSLSASKK103 pKa = 10.33ISQYY107 pKa = 11.25YY108 pKa = 7.22ITTITSFLNTGSMFFGVSGATLLNTPYY135 pKa = 10.09YY136 pKa = 9.93VKK138 pKa = 10.34SVYY141 pKa = 9.94SWSNTGMMVFKK152 pKa = 9.79RR153 pKa = 11.84TSGSAGSVVIHH164 pKa = 6.5LSVDD168 pKa = 3.21SSGVPTITNAGSICLYY184 pKa = 9.06NTDD187 pKa = 3.51WTQSTSIKK195 pKa = 10.46GSGCITVGPEE205 pKa = 3.78SQFNLEE211 pKa = 3.73MTAYY215 pKa = 9.87GISAAQTFYY224 pKa = 11.22LSSADD229 pKa = 3.51SVLSIHH235 pKa = 6.27GVVPGSSAYY244 pKa = 7.56PTYY247 pKa = 10.47RR248 pKa = 11.84VAGFGGGNTININLSFSKK266 pKa = 10.59FSYY269 pKa = 10.35SATTGVLTLSSALFTVYY286 pKa = 10.21FNIGTGYY293 pKa = 10.44KK294 pKa = 10.09SSLFSTNTAMMGRR307 pKa = 11.84SITYY311 pKa = 9.42DD312 pKa = 3.4GPPPNSAPAVCSCEE326 pKa = 3.49WSFPSVTTSLIASSTTAKK344 pKa = 10.26SAANSVTSASSSSSSSPVTEE364 pKa = 3.96STNVSEE370 pKa = 4.99LSSASASRR378 pKa = 11.84STVEE382 pKa = 4.01LSSSVVSDD390 pKa = 3.19SSVISTTTTVADD402 pKa = 4.0PFDD405 pKa = 4.01FSSFFEE411 pKa = 4.56KK412 pKa = 10.49SDD414 pKa = 3.61SSLTASLASASSPSAAVDD432 pKa = 4.33FSLTSSAIVASSLSSDD448 pKa = 3.43YY449 pKa = 11.58DD450 pKa = 3.65LASISTLTSASSSSANSYY468 pKa = 11.31SSLTSSSIVASSSSDD483 pKa = 3.02AFEE486 pKa = 4.79SPLTSSLATASSSSATFEE504 pKa = 4.46SSMSPSSIVASSSSAAFEE522 pKa = 4.7SPLSSSLATASSSSDD537 pKa = 2.95AFEE540 pKa = 4.83SPLTSSANVASSLSSDD556 pKa = 3.36SGLAATSSLASASSSSAGLSTSLSPSSIVASSSSAALEE594 pKa = 4.15SSLTSSLATASSSSAAFEE612 pKa = 4.52SSLSSNLATASSSSATFEE630 pKa = 4.46SSMSPSSIVSSSSSDD645 pKa = 3.1AFEE648 pKa = 4.83SPLTSSANVASSLSSDD664 pKa = 3.36SGLAATSSLASASSSSAGLSTSLSPSSIVASSSSAALEE702 pKa = 4.15SSLTSSLATASSSSAAFEE720 pKa = 4.52SSLSSNLATASSSSATFEE738 pKa = 4.46SSMSPSSIVSSSSSDD753 pKa = 3.1AFEE756 pKa = 4.83SPLTSSANVASSLSSDD772 pKa = 3.36SGLAATSSLASASSSSAGLSTSLSPSSIVASSSSAALEE810 pKa = 4.15SSLTSSLATASSSSAAFEE828 pKa = 4.52SSLSSNLATASSSSATFEE846 pKa = 4.46SSMSPSSIVSSSSSDD861 pKa = 3.1AFEE864 pKa = 4.83SPLTSSANVASSLSSDD880 pKa = 3.36SGLAATSSLASASSSSAGLSTSLSPSSIVASSSSAALEE918 pKa = 4.15SSLTSSLATASSSSAAFEE936 pKa = 4.37SSLTSSLATASSSSATFEE954 pKa = 4.46SSMSPSSIVSSSSSAAFEE972 pKa = 4.37SSLTSSLATASSSSAAFEE990 pKa = 4.54SSLSSSLATASGSSATFEE1008 pKa = 4.58SSMSPSSIVSSSSSAAFEE1026 pKa = 4.37SSLTSSLATASSSSAAFEE1044 pKa = 4.54SSLSSSLATASGSSATFEE1062 pKa = 4.58SSMSPSSIVASSSSAAFEE1080 pKa = 4.7SPLSSSLATASSSSDD1095 pKa = 2.95AFEE1098 pKa = 4.83SPLTSSANVASSLSSDD1114 pKa = 3.36SGLAATSSLASASSSSAGLSTSLSPSSIVASSSSADD1150 pKa = 3.21SSMLSAFDD1158 pKa = 3.77SSVTRR1163 pKa = 11.84DD1164 pKa = 3.59LTSSSSLAGLSLSSTSTSPVDD1185 pKa = 3.84FNSFITSSSGSTSGLSFAQKK1205 pKa = 9.05TSSGAISSLAMGSEE1219 pKa = 4.31SVQRR1223 pKa = 11.84FTTTLVSSKK1232 pKa = 10.73SGDD1235 pKa = 3.48SSASEE1240 pKa = 3.81PDD1242 pKa = 3.23AASVTRR1248 pKa = 11.84SLSSFVSGKK1257 pKa = 10.44SLDD1260 pKa = 3.75EE1261 pKa = 4.07NAISSSPGSVTDD1273 pKa = 4.04LPVISSSVFLGYY1285 pKa = 10.56HH1286 pKa = 5.62NSSVGMSSSSWNSMSSLAQGTKK1308 pKa = 9.1STVTSRR1314 pKa = 11.84TNSGGLSSTVSTGSMSADD1332 pKa = 2.97LVTASDD1338 pKa = 4.43FSLSATEE1345 pKa = 4.19TTSAGASASATQPGAGGTSAPLSSRR1370 pKa = 11.84TDD1372 pKa = 3.88FPVSSSSILGVGPSVVSISGHH1393 pKa = 4.8SAASIDD1399 pKa = 3.89VSATAAYY1406 pKa = 8.09PSTKK1410 pKa = 9.69ASSVIASGSSVVFGFASASGKK1431 pKa = 6.71TTASSNQFGSSALIEE1446 pKa = 4.42SATDD1450 pKa = 3.33AKK1452 pKa = 11.09NSIVLEE1458 pKa = 4.42TPALVNTATVTSEE1471 pKa = 3.56FDD1473 pKa = 2.92HH1474 pKa = 6.54YY1475 pKa = 11.52GAVSSDD1481 pKa = 3.27GASTSGGDD1489 pKa = 3.43VVVNKK1494 pKa = 9.86PADD1497 pKa = 4.09DD1498 pKa = 3.97YY1499 pKa = 11.84EE1500 pKa = 4.36EE1501 pKa = 5.09ASLTVSAISTDD1512 pKa = 3.46DD1513 pKa = 3.39VAVAEE1518 pKa = 4.46TQAPSTVHH1526 pKa = 6.63FSSQATRR1533 pKa = 11.84QNTWVSVSTGSSEE1546 pKa = 4.45SLSTASINIEE1556 pKa = 3.96NSAGSYY1562 pKa = 10.03NLSLAMVVLSVLALAFF1578 pKa = 4.14
MM1 pKa = 6.97KK2 pKa = 9.96TGIGALATAFMCLFGVISAVTITQNTVEE30 pKa = 4.34VSPISLSLGEE40 pKa = 5.13LYY42 pKa = 10.46INPGVYY48 pKa = 10.4YY49 pKa = 10.51SIVNNFQTLLARR61 pKa = 11.84SVYY64 pKa = 10.62NKK66 pKa = 10.43GGLFITAADD75 pKa = 4.07SYY77 pKa = 8.8TASVTMTGDD86 pKa = 2.85TFMNSGIVSFNSLSASKK103 pKa = 10.33ISQYY107 pKa = 11.25YY108 pKa = 7.22ITTITSFLNTGSMFFGVSGATLLNTPYY135 pKa = 10.09YY136 pKa = 9.93VKK138 pKa = 10.34SVYY141 pKa = 9.94SWSNTGMMVFKK152 pKa = 9.79RR153 pKa = 11.84TSGSAGSVVIHH164 pKa = 6.5LSVDD168 pKa = 3.21SSGVPTITNAGSICLYY184 pKa = 9.06NTDD187 pKa = 3.51WTQSTSIKK195 pKa = 10.46GSGCITVGPEE205 pKa = 3.78SQFNLEE211 pKa = 3.73MTAYY215 pKa = 9.87GISAAQTFYY224 pKa = 11.22LSSADD229 pKa = 3.51SVLSIHH235 pKa = 6.27GVVPGSSAYY244 pKa = 7.56PTYY247 pKa = 10.47RR248 pKa = 11.84VAGFGGGNTININLSFSKK266 pKa = 10.59FSYY269 pKa = 10.35SATTGVLTLSSALFTVYY286 pKa = 10.21FNIGTGYY293 pKa = 10.44KK294 pKa = 10.09SSLFSTNTAMMGRR307 pKa = 11.84SITYY311 pKa = 9.42DD312 pKa = 3.4GPPPNSAPAVCSCEE326 pKa = 3.49WSFPSVTTSLIASSTTAKK344 pKa = 10.26SAANSVTSASSSSSSSPVTEE364 pKa = 3.96STNVSEE370 pKa = 4.99LSSASASRR378 pKa = 11.84STVEE382 pKa = 4.01LSSSVVSDD390 pKa = 3.19SSVISTTTTVADD402 pKa = 4.0PFDD405 pKa = 4.01FSSFFEE411 pKa = 4.56KK412 pKa = 10.49SDD414 pKa = 3.61SSLTASLASASSPSAAVDD432 pKa = 4.33FSLTSSAIVASSLSSDD448 pKa = 3.43YY449 pKa = 11.58DD450 pKa = 3.65LASISTLTSASSSSANSYY468 pKa = 11.31SSLTSSSIVASSSSDD483 pKa = 3.02AFEE486 pKa = 4.79SPLTSSLATASSSSATFEE504 pKa = 4.46SSMSPSSIVASSSSAAFEE522 pKa = 4.7SPLSSSLATASSSSDD537 pKa = 2.95AFEE540 pKa = 4.83SPLTSSANVASSLSSDD556 pKa = 3.36SGLAATSSLASASSSSAGLSTSLSPSSIVASSSSAALEE594 pKa = 4.15SSLTSSLATASSSSAAFEE612 pKa = 4.52SSLSSNLATASSSSATFEE630 pKa = 4.46SSMSPSSIVSSSSSDD645 pKa = 3.1AFEE648 pKa = 4.83SPLTSSANVASSLSSDD664 pKa = 3.36SGLAATSSLASASSSSAGLSTSLSPSSIVASSSSAALEE702 pKa = 4.15SSLTSSLATASSSSAAFEE720 pKa = 4.52SSLSSNLATASSSSATFEE738 pKa = 4.46SSMSPSSIVSSSSSDD753 pKa = 3.1AFEE756 pKa = 4.83SPLTSSANVASSLSSDD772 pKa = 3.36SGLAATSSLASASSSSAGLSTSLSPSSIVASSSSAALEE810 pKa = 4.15SSLTSSLATASSSSAAFEE828 pKa = 4.52SSLSSNLATASSSSATFEE846 pKa = 4.46SSMSPSSIVSSSSSDD861 pKa = 3.1AFEE864 pKa = 4.83SPLTSSANVASSLSSDD880 pKa = 3.36SGLAATSSLASASSSSAGLSTSLSPSSIVASSSSAALEE918 pKa = 4.15SSLTSSLATASSSSAAFEE936 pKa = 4.37SSLTSSLATASSSSATFEE954 pKa = 4.46SSMSPSSIVSSSSSAAFEE972 pKa = 4.37SSLTSSLATASSSSAAFEE990 pKa = 4.54SSLSSSLATASGSSATFEE1008 pKa = 4.58SSMSPSSIVSSSSSAAFEE1026 pKa = 4.37SSLTSSLATASSSSAAFEE1044 pKa = 4.54SSLSSSLATASGSSATFEE1062 pKa = 4.58SSMSPSSIVASSSSAAFEE1080 pKa = 4.7SPLSSSLATASSSSDD1095 pKa = 2.95AFEE1098 pKa = 4.83SPLTSSANVASSLSSDD1114 pKa = 3.36SGLAATSSLASASSSSAGLSTSLSPSSIVASSSSADD1150 pKa = 3.21SSMLSAFDD1158 pKa = 3.77SSVTRR1163 pKa = 11.84DD1164 pKa = 3.59LTSSSSLAGLSLSSTSTSPVDD1185 pKa = 3.84FNSFITSSSGSTSGLSFAQKK1205 pKa = 9.05TSSGAISSLAMGSEE1219 pKa = 4.31SVQRR1223 pKa = 11.84FTTTLVSSKK1232 pKa = 10.73SGDD1235 pKa = 3.48SSASEE1240 pKa = 3.81PDD1242 pKa = 3.23AASVTRR1248 pKa = 11.84SLSSFVSGKK1257 pKa = 10.44SLDD1260 pKa = 3.75EE1261 pKa = 4.07NAISSSPGSVTDD1273 pKa = 4.04LPVISSSVFLGYY1285 pKa = 10.56HH1286 pKa = 5.62NSSVGMSSSSWNSMSSLAQGTKK1308 pKa = 9.1STVTSRR1314 pKa = 11.84TNSGGLSSTVSTGSMSADD1332 pKa = 2.97LVTASDD1338 pKa = 4.43FSLSATEE1345 pKa = 4.19TTSAGASASATQPGAGGTSAPLSSRR1370 pKa = 11.84TDD1372 pKa = 3.88FPVSSSSILGVGPSVVSISGHH1393 pKa = 4.8SAASIDD1399 pKa = 3.89VSATAAYY1406 pKa = 8.09PSTKK1410 pKa = 9.69ASSVIASGSSVVFGFASASGKK1431 pKa = 6.71TTASSNQFGSSALIEE1446 pKa = 4.42SATDD1450 pKa = 3.33AKK1452 pKa = 11.09NSIVLEE1458 pKa = 4.42TPALVNTATVTSEE1471 pKa = 3.56FDD1473 pKa = 2.92HH1474 pKa = 6.54YY1475 pKa = 11.52GAVSSDD1481 pKa = 3.27GASTSGGDD1489 pKa = 3.43VVVNKK1494 pKa = 9.86PADD1497 pKa = 4.09DD1498 pKa = 3.97YY1499 pKa = 11.84EE1500 pKa = 4.36EE1501 pKa = 5.09ASLTVSAISTDD1512 pKa = 3.46DD1513 pKa = 3.39VAVAEE1518 pKa = 4.46TQAPSTVHH1526 pKa = 6.63FSSQATRR1533 pKa = 11.84QNTWVSVSTGSSEE1546 pKa = 4.45SLSTASINIEE1556 pKa = 3.96NSAGSYY1562 pKa = 10.03NLSLAMVVLSVLALAFF1578 pKa = 4.14
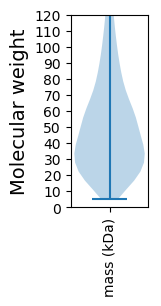
Molecular weight: 154.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P6XNB3|A0A4P6XNB3_9ASCO Ribosome biogenesis protein MAK21 OS=Metschnikowia aff. pulcherrima OX=2163413 GN=MPUL0C07910 PE=3 SV=1
MM1 pKa = 6.93PTEE4 pKa = 4.21TKK6 pKa = 10.03SGKK9 pKa = 9.84RR10 pKa = 11.84NPKK13 pKa = 9.05RR14 pKa = 11.84SLWRR18 pKa = 11.84NVRR21 pKa = 11.84CRR23 pKa = 11.84IAQSPYY29 pKa = 7.91IQQNKK34 pKa = 8.6SVSRR38 pKa = 11.84EE39 pKa = 3.43MRR41 pKa = 11.84RR42 pKa = 11.84FQRR45 pKa = 11.84EE46 pKa = 3.96NQDD49 pKa = 3.15KK50 pKa = 9.08RR51 pKa = 11.84TKK53 pKa = 9.95PQIACISRR61 pKa = 11.84YY62 pKa = 9.37RR63 pKa = 11.84QLKK66 pKa = 9.04PSTPYY71 pKa = 11.12GLVRR75 pKa = 11.84CLPIVFPNIPRR86 pKa = 11.84KK87 pKa = 9.41RR88 pKa = 11.84ARR90 pKa = 11.84ALATLGDD97 pKa = 3.88FRR99 pKa = 11.84IASPARR105 pKa = 11.84CHH107 pKa = 7.62LIFCCDD113 pKa = 3.94DD114 pKa = 3.44EE115 pKa = 5.03SAVIPGRR122 pKa = 11.84ARR124 pKa = 11.84TDD126 pKa = 3.21HH127 pKa = 6.4LCAYY131 pKa = 8.75QHH133 pKa = 6.23FAHH136 pKa = 6.86GNHH139 pKa = 5.54MHH141 pKa = 6.53TLHH144 pKa = 6.87SITYY148 pKa = 7.75TLL150 pKa = 3.84
MM1 pKa = 6.93PTEE4 pKa = 4.21TKK6 pKa = 10.03SGKK9 pKa = 9.84RR10 pKa = 11.84NPKK13 pKa = 9.05RR14 pKa = 11.84SLWRR18 pKa = 11.84NVRR21 pKa = 11.84CRR23 pKa = 11.84IAQSPYY29 pKa = 7.91IQQNKK34 pKa = 8.6SVSRR38 pKa = 11.84EE39 pKa = 3.43MRR41 pKa = 11.84RR42 pKa = 11.84FQRR45 pKa = 11.84EE46 pKa = 3.96NQDD49 pKa = 3.15KK50 pKa = 9.08RR51 pKa = 11.84TKK53 pKa = 9.95PQIACISRR61 pKa = 11.84YY62 pKa = 9.37RR63 pKa = 11.84QLKK66 pKa = 9.04PSTPYY71 pKa = 11.12GLVRR75 pKa = 11.84CLPIVFPNIPRR86 pKa = 11.84KK87 pKa = 9.41RR88 pKa = 11.84ARR90 pKa = 11.84ALATLGDD97 pKa = 3.88FRR99 pKa = 11.84IASPARR105 pKa = 11.84CHH107 pKa = 7.62LIFCCDD113 pKa = 3.94DD114 pKa = 3.44EE115 pKa = 5.03SAVIPGRR122 pKa = 11.84ARR124 pKa = 11.84TDD126 pKa = 3.21HH127 pKa = 6.4LCAYY131 pKa = 8.75QHH133 pKa = 6.23FAHH136 pKa = 6.86GNHH139 pKa = 5.54MHH141 pKa = 6.53TLHH144 pKa = 6.87SITYY148 pKa = 7.75TLL150 pKa = 3.84
Molecular weight: 17.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2902063 |
50 |
4996 |
500.8 |
56.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.357 ± 0.03 | 1.215 ± 0.012 |
5.698 ± 0.025 | 6.612 ± 0.032 |
4.457 ± 0.021 | 5.533 ± 0.028 |
2.407 ± 0.017 | 5.325 ± 0.025 |
6.328 ± 0.03 | 10.368 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.102 ± 0.013 | 4.594 ± 0.019 |
4.697 ± 0.029 | 3.868 ± 0.023 |
4.861 ± 0.025 | 8.365 ± 0.064 |
5.702 ± 0.057 | 6.349 ± 0.025 |
1.044 ± 0.011 | 3.12 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
