
Pararge aegeria rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Alphapaprhavirus; Pararge alphapaprhavirus
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
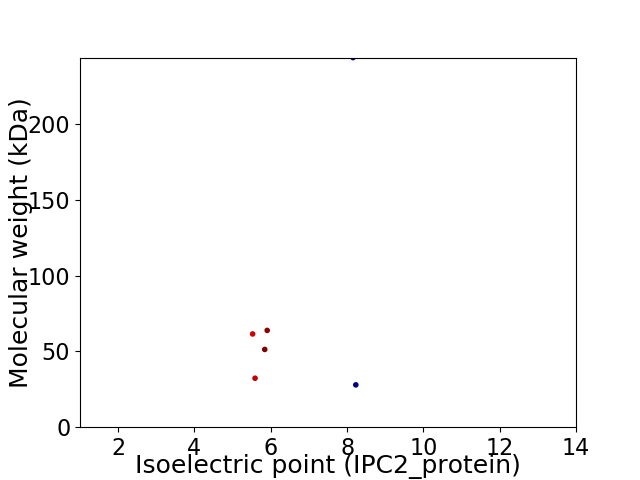
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140D8Q3|A0A140D8Q3_9RHAB GDP polyribonucleotidyltransferase OS=Pararge aegeria rhabdovirus OX=1802938 GN=L PE=4 SV=1
MM1 pKa = 7.45MICTLILIVVGFNNLPVSHH20 pKa = 7.0SSPRR24 pKa = 11.84DD25 pKa = 3.38FTSDD29 pKa = 3.26YY30 pKa = 11.26LLYY33 pKa = 9.2PVQMHH38 pKa = 6.32GSRR41 pKa = 11.84RR42 pKa = 11.84LASVDD47 pKa = 3.2RR48 pKa = 11.84LKK50 pKa = 11.02CPKK53 pKa = 10.51LSDD56 pKa = 3.56SLSKK60 pKa = 10.87VKK62 pKa = 10.7RR63 pKa = 11.84FDD65 pKa = 3.54LQLATAEE72 pKa = 3.82PRR74 pKa = 11.84TRR76 pKa = 11.84VGYY79 pKa = 10.22LCIGVTSDD87 pKa = 3.75IVCSSAILSYY97 pKa = 9.99ATSSSQKK104 pKa = 7.97TVHH107 pKa = 4.99VTKK110 pKa = 10.27SDD112 pKa = 3.73CEE114 pKa = 4.06VRR116 pKa = 11.84VSKK119 pKa = 10.76FSHH122 pKa = 5.68GQNTGFKK129 pKa = 8.7EE130 pKa = 4.32TTPDD134 pKa = 3.25CALGYY139 pKa = 9.34TSDD142 pKa = 4.26RR143 pKa = 11.84RR144 pKa = 11.84DD145 pKa = 3.13HH146 pKa = 6.31QIIVKK151 pKa = 9.84KK152 pKa = 10.53QIFPFSPLKK161 pKa = 10.71NGIVSSLFPGSVCADD176 pKa = 4.48RR177 pKa = 11.84YY178 pKa = 10.37CSSSDD183 pKa = 3.23GTTHH187 pKa = 7.3WFLEE191 pKa = 4.06SSGDD195 pKa = 3.75YY196 pKa = 11.09GCTGLVNGTGEE207 pKa = 4.48LKK209 pKa = 10.52ISTAGKK215 pKa = 10.25GRR217 pKa = 11.84SQLLSTSVTGDD228 pKa = 2.81IPLNEE233 pKa = 3.89ICKK236 pKa = 9.64TDD238 pKa = 4.08DD239 pKa = 3.3YY240 pKa = 11.41CGFPTYY246 pKa = 10.04IDD248 pKa = 3.49YY249 pKa = 11.36SFNGFSLTSLEE260 pKa = 3.94GDD262 pKa = 3.08IGSILAQDD270 pKa = 3.89VRR272 pKa = 11.84DD273 pKa = 4.07CPGGGSIRR281 pKa = 11.84EE282 pKa = 3.94YY283 pKa = 10.82HH284 pKa = 6.6PNDD287 pKa = 3.89LKK289 pKa = 10.66TMLSLTEE296 pKa = 4.64TIHH299 pKa = 5.6QRR301 pKa = 11.84HH302 pKa = 5.29YY303 pKa = 10.62QCQKK307 pKa = 9.74ILNKK311 pKa = 10.36AKK313 pKa = 10.66EE314 pKa = 4.17EE315 pKa = 4.28GFMSSRR321 pKa = 11.84YY322 pKa = 8.28LTLFQPYY329 pKa = 9.38QEE331 pKa = 5.38GIAPGYY337 pKa = 9.87RR338 pKa = 11.84LEE340 pKa = 4.74NGKK343 pKa = 9.34LWEE346 pKa = 4.08YY347 pKa = 11.2DD348 pKa = 3.13LYY350 pKa = 10.57YY351 pKa = 10.62TIGLFKK357 pKa = 10.83SQMIDD362 pKa = 2.79QTTYY366 pKa = 9.05KK367 pKa = 9.05TYY369 pKa = 10.07KK370 pKa = 9.01WDD372 pKa = 3.87VIDD375 pKa = 4.53ASGSFTTLPPSLCLFDD391 pKa = 3.55TTYY394 pKa = 11.18EE395 pKa = 4.01NSADD399 pKa = 3.74GPLCHH404 pKa = 6.94WFNGIQIHH412 pKa = 6.77GINLLYY418 pKa = 10.19PDD420 pKa = 3.4VANFFEE426 pKa = 4.53YY427 pKa = 10.64SHH429 pKa = 7.69DD430 pKa = 4.21YY431 pKa = 7.78PTLPYY436 pKa = 9.57STEE439 pKa = 4.12TTDD442 pKa = 4.12EE443 pKa = 4.29DD444 pKa = 4.35TDD446 pKa = 4.0LQHH449 pKa = 6.87NPTIPKK455 pKa = 9.53SWTGLLPFWGEE466 pKa = 3.97VLFLSISGVIFLLIIWLVLWSCCNGSKK493 pKa = 9.3TKK495 pKa = 10.6RR496 pKa = 11.84SGTEE500 pKa = 3.7TLNDD504 pKa = 3.83GGDD507 pKa = 3.59VKK509 pKa = 10.2ITEE512 pKa = 4.98LGTIRR517 pKa = 11.84PSLPQIRR524 pKa = 11.84QAIPLAWNQHH534 pKa = 5.48NIVTTRR540 pKa = 11.84HH541 pKa = 5.56SPPMLEE547 pKa = 4.16WFDD550 pKa = 3.65
MM1 pKa = 7.45MICTLILIVVGFNNLPVSHH20 pKa = 7.0SSPRR24 pKa = 11.84DD25 pKa = 3.38FTSDD29 pKa = 3.26YY30 pKa = 11.26LLYY33 pKa = 9.2PVQMHH38 pKa = 6.32GSRR41 pKa = 11.84RR42 pKa = 11.84LASVDD47 pKa = 3.2RR48 pKa = 11.84LKK50 pKa = 11.02CPKK53 pKa = 10.51LSDD56 pKa = 3.56SLSKK60 pKa = 10.87VKK62 pKa = 10.7RR63 pKa = 11.84FDD65 pKa = 3.54LQLATAEE72 pKa = 3.82PRR74 pKa = 11.84TRR76 pKa = 11.84VGYY79 pKa = 10.22LCIGVTSDD87 pKa = 3.75IVCSSAILSYY97 pKa = 9.99ATSSSQKK104 pKa = 7.97TVHH107 pKa = 4.99VTKK110 pKa = 10.27SDD112 pKa = 3.73CEE114 pKa = 4.06VRR116 pKa = 11.84VSKK119 pKa = 10.76FSHH122 pKa = 5.68GQNTGFKK129 pKa = 8.7EE130 pKa = 4.32TTPDD134 pKa = 3.25CALGYY139 pKa = 9.34TSDD142 pKa = 4.26RR143 pKa = 11.84RR144 pKa = 11.84DD145 pKa = 3.13HH146 pKa = 6.31QIIVKK151 pKa = 9.84KK152 pKa = 10.53QIFPFSPLKK161 pKa = 10.71NGIVSSLFPGSVCADD176 pKa = 4.48RR177 pKa = 11.84YY178 pKa = 10.37CSSSDD183 pKa = 3.23GTTHH187 pKa = 7.3WFLEE191 pKa = 4.06SSGDD195 pKa = 3.75YY196 pKa = 11.09GCTGLVNGTGEE207 pKa = 4.48LKK209 pKa = 10.52ISTAGKK215 pKa = 10.25GRR217 pKa = 11.84SQLLSTSVTGDD228 pKa = 2.81IPLNEE233 pKa = 3.89ICKK236 pKa = 9.64TDD238 pKa = 4.08DD239 pKa = 3.3YY240 pKa = 11.41CGFPTYY246 pKa = 10.04IDD248 pKa = 3.49YY249 pKa = 11.36SFNGFSLTSLEE260 pKa = 3.94GDD262 pKa = 3.08IGSILAQDD270 pKa = 3.89VRR272 pKa = 11.84DD273 pKa = 4.07CPGGGSIRR281 pKa = 11.84EE282 pKa = 3.94YY283 pKa = 10.82HH284 pKa = 6.6PNDD287 pKa = 3.89LKK289 pKa = 10.66TMLSLTEE296 pKa = 4.64TIHH299 pKa = 5.6QRR301 pKa = 11.84HH302 pKa = 5.29YY303 pKa = 10.62QCQKK307 pKa = 9.74ILNKK311 pKa = 10.36AKK313 pKa = 10.66EE314 pKa = 4.17EE315 pKa = 4.28GFMSSRR321 pKa = 11.84YY322 pKa = 8.28LTLFQPYY329 pKa = 9.38QEE331 pKa = 5.38GIAPGYY337 pKa = 9.87RR338 pKa = 11.84LEE340 pKa = 4.74NGKK343 pKa = 9.34LWEE346 pKa = 4.08YY347 pKa = 11.2DD348 pKa = 3.13LYY350 pKa = 10.57YY351 pKa = 10.62TIGLFKK357 pKa = 10.83SQMIDD362 pKa = 2.79QTTYY366 pKa = 9.05KK367 pKa = 9.05TYY369 pKa = 10.07KK370 pKa = 9.01WDD372 pKa = 3.87VIDD375 pKa = 4.53ASGSFTTLPPSLCLFDD391 pKa = 3.55TTYY394 pKa = 11.18EE395 pKa = 4.01NSADD399 pKa = 3.74GPLCHH404 pKa = 6.94WFNGIQIHH412 pKa = 6.77GINLLYY418 pKa = 10.19PDD420 pKa = 3.4VANFFEE426 pKa = 4.53YY427 pKa = 10.64SHH429 pKa = 7.69DD430 pKa = 4.21YY431 pKa = 7.78PTLPYY436 pKa = 9.57STEE439 pKa = 4.12TTDD442 pKa = 4.12EE443 pKa = 4.29DD444 pKa = 4.35TDD446 pKa = 4.0LQHH449 pKa = 6.87NPTIPKK455 pKa = 9.53SWTGLLPFWGEE466 pKa = 3.97VLFLSISGVIFLLIIWLVLWSCCNGSKK493 pKa = 9.3TKK495 pKa = 10.6RR496 pKa = 11.84SGTEE500 pKa = 3.7TLNDD504 pKa = 3.83GGDD507 pKa = 3.59VKK509 pKa = 10.2ITEE512 pKa = 4.98LGTIRR517 pKa = 11.84PSLPQIRR524 pKa = 11.84QAIPLAWNQHH534 pKa = 5.48NIVTTRR540 pKa = 11.84HH541 pKa = 5.56SPPMLEE547 pKa = 4.16WFDD550 pKa = 3.65
Molecular weight: 61.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140D8Q1|A0A140D8Q1_9RHAB Glycoprotein 1 OS=Pararge aegeria rhabdovirus OX=1802938 GN=G1 PE=4 SV=1
MM1 pKa = 7.47SSSTSSSSSEE11 pKa = 3.9TKK13 pKa = 8.62KK14 pKa = 10.69QKK16 pKa = 9.48VRR18 pKa = 11.84RR19 pKa = 11.84QRR21 pKa = 11.84TFLGIRR27 pKa = 11.84WPRR30 pKa = 11.84QSSSSDD36 pKa = 3.15SLSPSEE42 pKa = 4.28GLNPLALPGRR52 pKa = 11.84SSGPTIIPSGFMMGGSVNWANPEE75 pKa = 3.59WDD77 pKa = 3.39FHH79 pKa = 7.9KK80 pKa = 11.07EE81 pKa = 3.83EE82 pKa = 4.24VSDD85 pKa = 4.0VVTILPFPCQVTCTLMIVSRR105 pKa = 11.84KK106 pKa = 9.23PISDD110 pKa = 3.62VQSLLRR116 pKa = 11.84KK117 pKa = 10.04LEE119 pKa = 3.99LWVDD123 pKa = 3.92QYY125 pKa = 11.77SGSYY129 pKa = 9.65VSRR132 pKa = 11.84PYY134 pKa = 9.85LTLIYY139 pKa = 10.0ILLGAKK145 pKa = 8.02LTSRR149 pKa = 11.84KK150 pKa = 8.41STSRR154 pKa = 11.84NYY156 pKa = 9.94EE157 pKa = 3.83YY158 pKa = 10.38HH159 pKa = 7.13SKK161 pKa = 9.63FQGVIRR167 pKa = 11.84FGIQSDD173 pKa = 3.98RR174 pKa = 11.84NKK176 pKa = 10.42PPAEE180 pKa = 3.93NFRR183 pKa = 11.84LVMSEE188 pKa = 3.92GLEE191 pKa = 3.84DD192 pKa = 3.14SYY194 pKa = 11.86YY195 pKa = 10.8AISFEE200 pKa = 4.84CYY202 pKa = 9.67SSEE205 pKa = 4.18SKK207 pKa = 10.61RR208 pKa = 11.84KK209 pKa = 9.14GVIYY213 pKa = 10.6SDD215 pKa = 4.06LYY217 pKa = 11.06KK218 pKa = 10.5MNGYY222 pKa = 10.27HH223 pKa = 5.28YY224 pKa = 7.96TFPCAMEE231 pKa = 4.41LLGLQVVMDD240 pKa = 3.99QEE242 pKa = 4.45GLIASAA248 pKa = 4.71
MM1 pKa = 7.47SSSTSSSSSEE11 pKa = 3.9TKK13 pKa = 8.62KK14 pKa = 10.69QKK16 pKa = 9.48VRR18 pKa = 11.84RR19 pKa = 11.84QRR21 pKa = 11.84TFLGIRR27 pKa = 11.84WPRR30 pKa = 11.84QSSSSDD36 pKa = 3.15SLSPSEE42 pKa = 4.28GLNPLALPGRR52 pKa = 11.84SSGPTIIPSGFMMGGSVNWANPEE75 pKa = 3.59WDD77 pKa = 3.39FHH79 pKa = 7.9KK80 pKa = 11.07EE81 pKa = 3.83EE82 pKa = 4.24VSDD85 pKa = 4.0VVTILPFPCQVTCTLMIVSRR105 pKa = 11.84KK106 pKa = 9.23PISDD110 pKa = 3.62VQSLLRR116 pKa = 11.84KK117 pKa = 10.04LEE119 pKa = 3.99LWVDD123 pKa = 3.92QYY125 pKa = 11.77SGSYY129 pKa = 9.65VSRR132 pKa = 11.84PYY134 pKa = 9.85LTLIYY139 pKa = 10.0ILLGAKK145 pKa = 8.02LTSRR149 pKa = 11.84KK150 pKa = 8.41STSRR154 pKa = 11.84NYY156 pKa = 9.94EE157 pKa = 3.83YY158 pKa = 10.38HH159 pKa = 7.13SKK161 pKa = 9.63FQGVIRR167 pKa = 11.84FGIQSDD173 pKa = 3.98RR174 pKa = 11.84NKK176 pKa = 10.42PPAEE180 pKa = 3.93NFRR183 pKa = 11.84LVMSEE188 pKa = 3.92GLEE191 pKa = 3.84DD192 pKa = 3.14SYY194 pKa = 11.86YY195 pKa = 10.8AISFEE200 pKa = 4.84CYY202 pKa = 9.67SSEE205 pKa = 4.18SKK207 pKa = 10.61RR208 pKa = 11.84KK209 pKa = 9.14GVIYY213 pKa = 10.6SDD215 pKa = 4.06LYY217 pKa = 11.06KK218 pKa = 10.5MNGYY222 pKa = 10.27HH223 pKa = 5.28YY224 pKa = 7.96TFPCAMEE231 pKa = 4.41LLGLQVVMDD240 pKa = 3.99QEE242 pKa = 4.45GLIASAA248 pKa = 4.71
Molecular weight: 27.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4246 |
248 |
2135 |
707.7 |
80.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.91 ± 0.505 | 2.002 ± 0.313 |
5.417 ± 0.343 | 5.37 ± 0.369 |
3.745 ± 0.228 | 7.065 ± 0.345 |
2.591 ± 0.176 | 7.042 ± 0.547 |
5.582 ± 0.297 | 11.187 ± 0.83 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.214 ± 0.302 | 4.31 ± 0.308 |
5.252 ± 0.171 | 3.58 ± 0.164 |
5.535 ± 0.391 | 8.455 ± 0.893 |
5.888 ± 0.545 | 5.134 ± 0.268 |
1.743 ± 0.092 | 3.98 ± 0.304 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
