
Flavobacterium psychrotolerans
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
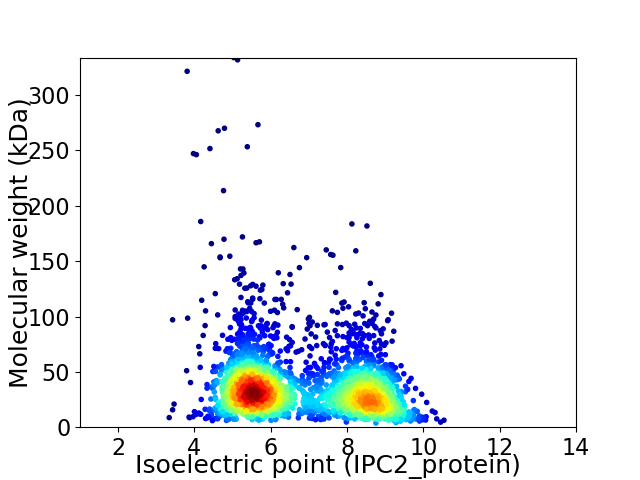
Virtual 2D-PAGE plot for 2753 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U1JMW5|A0A2U1JMW5_9FLAO Urease accessory protein UreE OS=Flavobacterium psychrotolerans OX=2169410 GN=ureE PE=3 SV=1
MM1 pKa = 7.51KK2 pKa = 10.42KK3 pKa = 8.47ITLLFMLFIGFSAFAQFPQDD23 pKa = 3.54FEE25 pKa = 4.86GTFPPTGWAVYY36 pKa = 10.08RR37 pKa = 11.84GANDD41 pKa = 4.43LGIEE45 pKa = 4.22SDD47 pKa = 3.4WTATTVGVATGLQAAYY63 pKa = 10.5VDD65 pKa = 4.39YY66 pKa = 10.8EE67 pKa = 4.13AVTAGEE73 pKa = 4.26LAEE76 pKa = 4.79DD77 pKa = 3.9WLVSPAYY84 pKa = 9.28TVTAPNTLMAFYY96 pKa = 10.11QGQDD100 pKa = 3.12DD101 pKa = 4.03PTDD104 pKa = 3.52YY105 pKa = 11.36GSTYY109 pKa = 9.27TIRR112 pKa = 11.84VSTTSQIDD120 pKa = 3.4INSFTIVDD128 pKa = 4.01TQTEE132 pKa = 4.34SDD134 pKa = 3.75FTIEE138 pKa = 3.9MTTRR142 pKa = 11.84TVDD145 pKa = 3.51LSAYY149 pKa = 9.63AGQTIYY155 pKa = 10.61IAFVLEE161 pKa = 4.15NNEE164 pKa = 3.58GDD166 pKa = 3.06AWAIDD171 pKa = 4.43GIDD174 pKa = 4.05FISNTVIAPGCVSNPSPADD193 pKa = 3.12AALEE197 pKa = 4.21VPIGATTFSWTAPTTGGPVTLYY219 pKa = 10.64NIYY222 pKa = 10.73SGATSDD228 pKa = 4.45AVTDD232 pKa = 5.27FITSTTTPSVDD243 pKa = 3.54LSLADD248 pKa = 4.71FSTTFYY254 pKa = 10.18WKK256 pKa = 10.05VVAVNLVGSATDD268 pKa = 3.46CAVWSFTTQSPPGYY282 pKa = 9.57CLAASGGQYY291 pKa = 10.05PADD294 pKa = 3.97AYY296 pKa = 9.99TPEE299 pKa = 4.52ILDD302 pKa = 3.76GVTEE306 pKa = 4.41NIIVSDD312 pKa = 4.19GFAGEE317 pKa = 4.04YY318 pKa = 10.45SVVNVLSGQTYY329 pKa = 7.8TFTSGTTDD337 pKa = 4.9FITIGNEE344 pKa = 3.76AGDD347 pKa = 3.67TSLIAGASPLTWVSDD362 pKa = 3.13ISGVIRR368 pKa = 11.84FYY370 pKa = 11.61SHH372 pKa = 7.32VDD374 pKa = 3.48DD375 pKa = 4.26QCGEE379 pKa = 4.12EE380 pKa = 4.32NIEE383 pKa = 4.07RR384 pKa = 11.84PRR386 pKa = 11.84TVISDD391 pKa = 3.86PLLATDD397 pKa = 4.73TFNSASFHH405 pKa = 5.7TFPNPVKK412 pKa = 10.59DD413 pKa = 3.88ILNISYY419 pKa = 10.42DD420 pKa = 3.76KK421 pKa = 11.01NISNVAVFNLLGQEE435 pKa = 4.16VMTKK439 pKa = 8.87SVKK442 pKa = 9.96QNQSQVDD449 pKa = 3.91MSHH452 pKa = 6.92LSNGTYY458 pKa = 8.32MVKK461 pKa = 8.8VTSDD465 pKa = 3.46NQVKK469 pKa = 8.88TIKK472 pKa = 10.31VIKK475 pKa = 10.09GG476 pKa = 3.21
MM1 pKa = 7.51KK2 pKa = 10.42KK3 pKa = 8.47ITLLFMLFIGFSAFAQFPQDD23 pKa = 3.54FEE25 pKa = 4.86GTFPPTGWAVYY36 pKa = 10.08RR37 pKa = 11.84GANDD41 pKa = 4.43LGIEE45 pKa = 4.22SDD47 pKa = 3.4WTATTVGVATGLQAAYY63 pKa = 10.5VDD65 pKa = 4.39YY66 pKa = 10.8EE67 pKa = 4.13AVTAGEE73 pKa = 4.26LAEE76 pKa = 4.79DD77 pKa = 3.9WLVSPAYY84 pKa = 9.28TVTAPNTLMAFYY96 pKa = 10.11QGQDD100 pKa = 3.12DD101 pKa = 4.03PTDD104 pKa = 3.52YY105 pKa = 11.36GSTYY109 pKa = 9.27TIRR112 pKa = 11.84VSTTSQIDD120 pKa = 3.4INSFTIVDD128 pKa = 4.01TQTEE132 pKa = 4.34SDD134 pKa = 3.75FTIEE138 pKa = 3.9MTTRR142 pKa = 11.84TVDD145 pKa = 3.51LSAYY149 pKa = 9.63AGQTIYY155 pKa = 10.61IAFVLEE161 pKa = 4.15NNEE164 pKa = 3.58GDD166 pKa = 3.06AWAIDD171 pKa = 4.43GIDD174 pKa = 4.05FISNTVIAPGCVSNPSPADD193 pKa = 3.12AALEE197 pKa = 4.21VPIGATTFSWTAPTTGGPVTLYY219 pKa = 10.64NIYY222 pKa = 10.73SGATSDD228 pKa = 4.45AVTDD232 pKa = 5.27FITSTTTPSVDD243 pKa = 3.54LSLADD248 pKa = 4.71FSTTFYY254 pKa = 10.18WKK256 pKa = 10.05VVAVNLVGSATDD268 pKa = 3.46CAVWSFTTQSPPGYY282 pKa = 9.57CLAASGGQYY291 pKa = 10.05PADD294 pKa = 3.97AYY296 pKa = 9.99TPEE299 pKa = 4.52ILDD302 pKa = 3.76GVTEE306 pKa = 4.41NIIVSDD312 pKa = 4.19GFAGEE317 pKa = 4.04YY318 pKa = 10.45SVVNVLSGQTYY329 pKa = 7.8TFTSGTTDD337 pKa = 4.9FITIGNEE344 pKa = 3.76AGDD347 pKa = 3.67TSLIAGASPLTWVSDD362 pKa = 3.13ISGVIRR368 pKa = 11.84FYY370 pKa = 11.61SHH372 pKa = 7.32VDD374 pKa = 3.48DD375 pKa = 4.26QCGEE379 pKa = 4.12EE380 pKa = 4.32NIEE383 pKa = 4.07RR384 pKa = 11.84PRR386 pKa = 11.84TVISDD391 pKa = 3.86PLLATDD397 pKa = 4.73TFNSASFHH405 pKa = 5.7TFPNPVKK412 pKa = 10.59DD413 pKa = 3.88ILNISYY419 pKa = 10.42DD420 pKa = 3.76KK421 pKa = 11.01NISNVAVFNLLGQEE435 pKa = 4.16VMTKK439 pKa = 8.87SVKK442 pKa = 9.96QNQSQVDD449 pKa = 3.91MSHH452 pKa = 6.92LSNGTYY458 pKa = 8.32MVKK461 pKa = 8.8VTSDD465 pKa = 3.46NQVKK469 pKa = 8.88TIKK472 pKa = 10.31VIKK475 pKa = 10.09GG476 pKa = 3.21
Molecular weight: 51.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U1JFJ3|A0A2U1JFJ3_9FLAO IS91 family transposase OS=Flavobacterium psychrotolerans OX=2169410 GN=DB895_14025 PE=4 SV=1
RR1 pKa = 7.43GAPGVDD7 pKa = 2.48RR8 pKa = 11.84LTVEE12 pKa = 4.37EE13 pKa = 4.4LKK15 pKa = 10.93DD16 pKa = 3.23WLLVHH21 pKa = 6.55WPSVKK26 pKa = 10.32AALLEE31 pKa = 4.08DD32 pKa = 4.47RR33 pKa = 11.84YY34 pKa = 10.91LPRR37 pKa = 11.84AVRR40 pKa = 11.84RR41 pKa = 11.84VDD43 pKa = 3.0IPKK46 pKa = 9.93PNGGVRR52 pKa = 11.84TLGVPTVVDD61 pKa = 3.65RR62 pKa = 11.84LIQQALHH69 pKa = 5.99QVLQPIFEE77 pKa = 4.52PTFSDD82 pKa = 3.39GSYY85 pKa = 10.61GFRR88 pKa = 11.84PQRR91 pKa = 11.84SALDD95 pKa = 3.4AVVKK99 pKa = 10.48AKK101 pKa = 10.59EE102 pKa = 3.82YY103 pKa = 9.61VAEE106 pKa = 4.68GKK108 pKa = 9.58HH109 pKa = 4.47WVVDD113 pKa = 3.15IDD115 pKa = 3.99LEE117 pKa = 4.57KK118 pKa = 11.04FFDD121 pKa = 3.98RR122 pKa = 11.84VNHH125 pKa = 6.24DD126 pKa = 3.15VLMARR131 pKa = 11.84LAYY134 pKa = 9.58RR135 pKa = 11.84VRR137 pKa = 11.84DD138 pKa = 3.62GRR140 pKa = 11.84VLKK143 pKa = 10.51LIRR146 pKa = 11.84RR147 pKa = 11.84FLEE150 pKa = 4.05AGMMANGVIQPRR162 pKa = 11.84SEE164 pKa = 3.88GTPQGGPLSPLLSNILLTDD183 pKa = 4.08LDD185 pKa = 4.16RR186 pKa = 11.84EE187 pKa = 4.33LEE189 pKa = 3.97RR190 pKa = 11.84RR191 pKa = 11.84QLKK194 pKa = 8.83FCRR197 pKa = 11.84YY198 pKa = 10.23ADD200 pKa = 3.8DD201 pKa = 4.04CNIYY205 pKa = 10.13IGSEE209 pKa = 4.15GAGQRR214 pKa = 11.84AMANIRR220 pKa = 11.84AFLEE224 pKa = 4.02GHH226 pKa = 6.27LKK228 pKa = 10.61LRR230 pKa = 11.84INEE233 pKa = 4.05QKK235 pKa = 10.77SAVARR240 pKa = 11.84PWKK243 pKa = 10.37RR244 pKa = 11.84KK245 pKa = 8.5FLGYY249 pKa = 10.52AITIYY254 pKa = 10.3RR255 pKa = 11.84QEE257 pKa = 4.05TRR259 pKa = 11.84VRR261 pKa = 11.84PAPEE265 pKa = 3.66SLRR268 pKa = 11.84RR269 pKa = 11.84LMDD272 pKa = 3.49RR273 pKa = 11.84VRR275 pKa = 11.84EE276 pKa = 3.97LLRR279 pKa = 11.84KK280 pKa = 9.72GRR282 pKa = 11.84GRR284 pKa = 11.84SLSHH288 pKa = 6.64TIEE291 pKa = 3.9MLNPVLRR298 pKa = 11.84GWANYY303 pKa = 8.69FRR305 pKa = 11.84LTANMRR311 pKa = 11.84MLEE314 pKa = 3.99EE315 pKa = 4.4LDD317 pKa = 2.56WWLRR321 pKa = 11.84RR322 pKa = 11.84KK323 pKa = 9.23LRR325 pKa = 11.84CLLWRR330 pKa = 11.84QWKK333 pKa = 7.62TPKK336 pKa = 9.11TRR338 pKa = 11.84EE339 pKa = 3.55RR340 pKa = 11.84RR341 pKa = 11.84LILLGLRR348 pKa = 11.84PEE350 pKa = 4.41RR351 pKa = 11.84AWKK354 pKa = 10.52SSVNGRR360 pKa = 11.84GPWWNSASKK369 pKa = 10.6HH370 pKa = 4.88MNQAVPTLYY379 pKa = 10.88
RR1 pKa = 7.43GAPGVDD7 pKa = 2.48RR8 pKa = 11.84LTVEE12 pKa = 4.37EE13 pKa = 4.4LKK15 pKa = 10.93DD16 pKa = 3.23WLLVHH21 pKa = 6.55WPSVKK26 pKa = 10.32AALLEE31 pKa = 4.08DD32 pKa = 4.47RR33 pKa = 11.84YY34 pKa = 10.91LPRR37 pKa = 11.84AVRR40 pKa = 11.84RR41 pKa = 11.84VDD43 pKa = 3.0IPKK46 pKa = 9.93PNGGVRR52 pKa = 11.84TLGVPTVVDD61 pKa = 3.65RR62 pKa = 11.84LIQQALHH69 pKa = 5.99QVLQPIFEE77 pKa = 4.52PTFSDD82 pKa = 3.39GSYY85 pKa = 10.61GFRR88 pKa = 11.84PQRR91 pKa = 11.84SALDD95 pKa = 3.4AVVKK99 pKa = 10.48AKK101 pKa = 10.59EE102 pKa = 3.82YY103 pKa = 9.61VAEE106 pKa = 4.68GKK108 pKa = 9.58HH109 pKa = 4.47WVVDD113 pKa = 3.15IDD115 pKa = 3.99LEE117 pKa = 4.57KK118 pKa = 11.04FFDD121 pKa = 3.98RR122 pKa = 11.84VNHH125 pKa = 6.24DD126 pKa = 3.15VLMARR131 pKa = 11.84LAYY134 pKa = 9.58RR135 pKa = 11.84VRR137 pKa = 11.84DD138 pKa = 3.62GRR140 pKa = 11.84VLKK143 pKa = 10.51LIRR146 pKa = 11.84RR147 pKa = 11.84FLEE150 pKa = 4.05AGMMANGVIQPRR162 pKa = 11.84SEE164 pKa = 3.88GTPQGGPLSPLLSNILLTDD183 pKa = 4.08LDD185 pKa = 4.16RR186 pKa = 11.84EE187 pKa = 4.33LEE189 pKa = 3.97RR190 pKa = 11.84RR191 pKa = 11.84QLKK194 pKa = 8.83FCRR197 pKa = 11.84YY198 pKa = 10.23ADD200 pKa = 3.8DD201 pKa = 4.04CNIYY205 pKa = 10.13IGSEE209 pKa = 4.15GAGQRR214 pKa = 11.84AMANIRR220 pKa = 11.84AFLEE224 pKa = 4.02GHH226 pKa = 6.27LKK228 pKa = 10.61LRR230 pKa = 11.84INEE233 pKa = 4.05QKK235 pKa = 10.77SAVARR240 pKa = 11.84PWKK243 pKa = 10.37RR244 pKa = 11.84KK245 pKa = 8.5FLGYY249 pKa = 10.52AITIYY254 pKa = 10.3RR255 pKa = 11.84QEE257 pKa = 4.05TRR259 pKa = 11.84VRR261 pKa = 11.84PAPEE265 pKa = 3.66SLRR268 pKa = 11.84RR269 pKa = 11.84LMDD272 pKa = 3.49RR273 pKa = 11.84VRR275 pKa = 11.84EE276 pKa = 3.97LLRR279 pKa = 11.84KK280 pKa = 9.72GRR282 pKa = 11.84GRR284 pKa = 11.84SLSHH288 pKa = 6.64TIEE291 pKa = 3.9MLNPVLRR298 pKa = 11.84GWANYY303 pKa = 8.69FRR305 pKa = 11.84LTANMRR311 pKa = 11.84MLEE314 pKa = 3.99EE315 pKa = 4.4LDD317 pKa = 2.56WWLRR321 pKa = 11.84RR322 pKa = 11.84KK323 pKa = 9.23LRR325 pKa = 11.84CLLWRR330 pKa = 11.84QWKK333 pKa = 7.62TPKK336 pKa = 9.11TRR338 pKa = 11.84EE339 pKa = 3.55RR340 pKa = 11.84RR341 pKa = 11.84LILLGLRR348 pKa = 11.84PEE350 pKa = 4.41RR351 pKa = 11.84AWKK354 pKa = 10.52SSVNGRR360 pKa = 11.84GPWWNSASKK369 pKa = 10.6HH370 pKa = 4.88MNQAVPTLYY379 pKa = 10.88
Molecular weight: 44.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
953119 |
34 |
3340 |
346.2 |
38.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.433 ± 0.061 | 0.824 ± 0.016 |
5.124 ± 0.036 | 6.068 ± 0.071 |
5.385 ± 0.044 | 6.521 ± 0.062 |
1.689 ± 0.025 | 8.414 ± 0.044 |
7.886 ± 0.072 | 9.129 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.223 ± 0.025 | 6.398 ± 0.049 |
3.4 ± 0.027 | 3.346 ± 0.025 |
3.061 ± 0.03 | 6.679 ± 0.052 |
6.251 ± 0.103 | 6.254 ± 0.037 |
1.003 ± 0.018 | 3.911 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
