
Candidatus Hepatoplasma crinochetorum Av
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Candidatus Hepatoplasma; Candidatus Hepatoplasma crinochetorum
Average proteome isoelectric point is 7.92
Get precalculated fractions of proteins
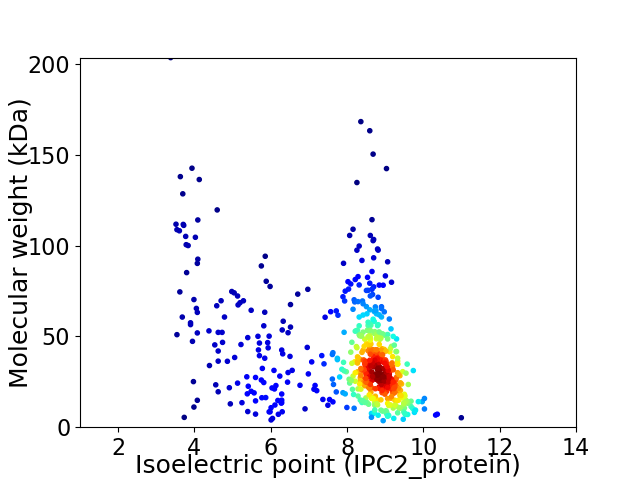
Virtual 2D-PAGE plot for 582 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
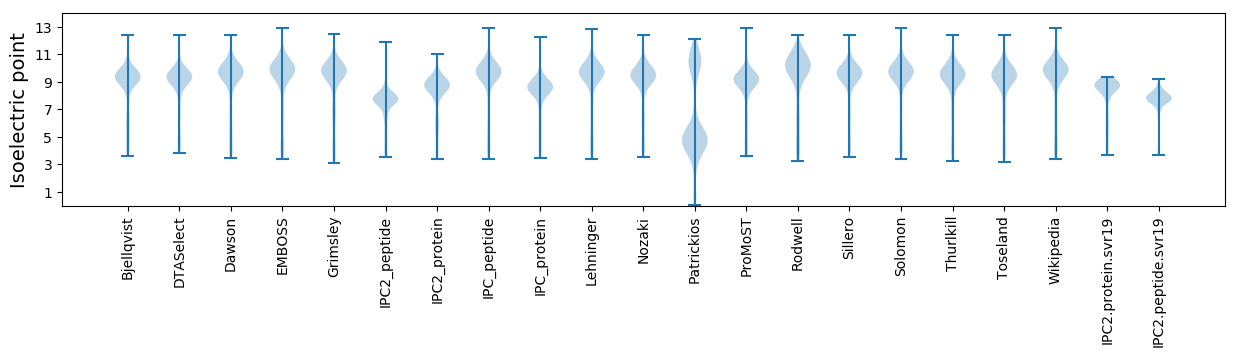
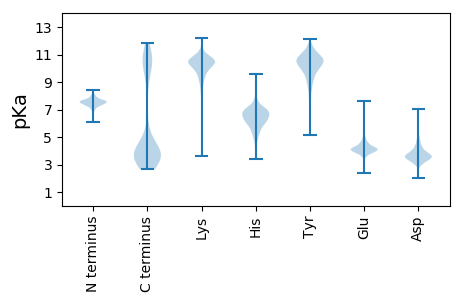
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8GMW0|W8GMW0_9MOLU Isoleucine--tRNA ligase OS=Candidatus Hepatoplasma crinochetorum Av OX=1427984 GN=ileS PE=3 SV=1
MM1 pKa = 7.97IDD3 pKa = 3.27GCNGGNIDD11 pKa = 4.07ILSINTINYY20 pKa = 8.81NSEE23 pKa = 3.91GEE25 pKa = 4.25KK26 pKa = 8.61TQKK29 pKa = 9.98EE30 pKa = 4.26VNEE33 pKa = 4.37TNKK36 pKa = 10.38EE37 pKa = 4.14AIDD40 pKa = 4.06DD41 pKa = 4.85LINNNMVDD49 pKa = 4.05TDD51 pKa = 3.8HH52 pKa = 6.19VTYY55 pKa = 9.91TVLGNTSTEE64 pKa = 3.81AFGDD68 pKa = 4.05FTQADD73 pKa = 3.74EE74 pKa = 4.3KK75 pKa = 11.41LYY77 pKa = 11.33GEE79 pKa = 4.48STNDD83 pKa = 3.09TVVRR87 pKa = 11.84DD88 pKa = 3.81NTLLGLTDD96 pKa = 4.48GLQDD100 pKa = 4.95QIDD103 pKa = 4.19GLNTAVNNLGDD114 pKa = 4.57IINNAIGDD122 pKa = 4.01IKK124 pKa = 11.16ASLDD128 pKa = 3.64SLSTCKK134 pKa = 10.5DD135 pKa = 3.35CSIDD139 pKa = 3.76PFPSAIIDD147 pKa = 3.97NIVPTDD153 pKa = 3.59PTTEE157 pKa = 4.45DD158 pKa = 3.44GNGTVSFDD166 pKa = 4.65VYY168 pKa = 11.26LSDD171 pKa = 5.45CNGTSIAVLNPASGTEE187 pKa = 4.32PITLSEE193 pKa = 4.71GINTGLTFTDD203 pKa = 5.1LPPGYY208 pKa = 10.54YY209 pKa = 10.17NIEE212 pKa = 4.32VYY214 pKa = 10.73CDD216 pKa = 3.26TEE218 pKa = 5.84LIDD221 pKa = 3.54NTSFIINDD229 pKa = 3.52YY230 pKa = 11.06DD231 pKa = 3.92SSEE234 pKa = 4.51DD235 pKa = 3.44GGGDD239 pKa = 3.54EE240 pKa = 5.03EE241 pKa = 6.1IIDD244 pKa = 4.46IVTPDD249 pKa = 4.78DD250 pKa = 4.27GSLIPPNSNDD260 pKa = 2.89IPVNDD265 pKa = 4.51GEE267 pKa = 4.66DD268 pKa = 3.64FNHH271 pKa = 6.84ILMITKK277 pKa = 10.4GNLIPEE283 pKa = 4.35NNIQCYY289 pKa = 9.59QNLFIVGSDD298 pKa = 3.46FNLNEE303 pKa = 3.95WTTGCLNSFNPNDD316 pKa = 3.9PQGDD320 pKa = 3.59EE321 pKa = 4.73CYY323 pKa = 10.68VAIKK327 pKa = 9.35RR328 pKa = 11.84TSDD331 pKa = 3.03NTFDD335 pKa = 3.54TTRR338 pKa = 11.84EE339 pKa = 4.16SVSNSVKK346 pKa = 10.02GLNWIKK352 pKa = 10.78AIDD355 pKa = 4.41NITITKK361 pKa = 9.68ILEE364 pKa = 4.24NGAFGYY370 pKa = 10.7GDD372 pKa = 3.48KK373 pKa = 10.83FVLNEE378 pKa = 4.62DD379 pKa = 3.56YY380 pKa = 11.18TNYY383 pKa = 11.11DD384 pKa = 3.94LIISKK389 pKa = 9.54WYY391 pKa = 9.93FNTSAGTAVKK401 pKa = 9.12PQYY404 pKa = 10.29GYY406 pKa = 9.78PIRR409 pKa = 11.84EE410 pKa = 3.91ISDD413 pKa = 4.77LIVTNTWLQKK423 pKa = 8.94QHH425 pKa = 5.66GSNVSYY431 pKa = 10.7SGEE434 pKa = 3.97LTFTDD439 pKa = 4.02TLNGTVTKK447 pKa = 10.61ARR449 pKa = 11.84EE450 pKa = 4.04SDD452 pKa = 3.99FLATMWGVNGIPAPIDD468 pKa = 3.42LNAQVSGTIINMDD481 pKa = 4.28DD482 pKa = 3.55YY483 pKa = 12.02SNADD487 pKa = 3.55FLLINCTYY495 pKa = 11.37ADD497 pKa = 3.47IGNTQLMYY505 pKa = 11.0AKK507 pKa = 10.3DD508 pKa = 3.31IVYY511 pKa = 10.45DD512 pKa = 3.32GTTRR516 pKa = 11.84YY517 pKa = 10.84DD518 pKa = 3.33FVGIGACYY526 pKa = 10.12FSFNVDD532 pKa = 2.88KK533 pKa = 10.53TINIIDD539 pKa = 3.93AALSAEE545 pKa = 4.45IIITSIKK552 pKa = 10.36KK553 pKa = 10.22IILL556 pKa = 3.78
MM1 pKa = 7.97IDD3 pKa = 3.27GCNGGNIDD11 pKa = 4.07ILSINTINYY20 pKa = 8.81NSEE23 pKa = 3.91GEE25 pKa = 4.25KK26 pKa = 8.61TQKK29 pKa = 9.98EE30 pKa = 4.26VNEE33 pKa = 4.37TNKK36 pKa = 10.38EE37 pKa = 4.14AIDD40 pKa = 4.06DD41 pKa = 4.85LINNNMVDD49 pKa = 4.05TDD51 pKa = 3.8HH52 pKa = 6.19VTYY55 pKa = 9.91TVLGNTSTEE64 pKa = 3.81AFGDD68 pKa = 4.05FTQADD73 pKa = 3.74EE74 pKa = 4.3KK75 pKa = 11.41LYY77 pKa = 11.33GEE79 pKa = 4.48STNDD83 pKa = 3.09TVVRR87 pKa = 11.84DD88 pKa = 3.81NTLLGLTDD96 pKa = 4.48GLQDD100 pKa = 4.95QIDD103 pKa = 4.19GLNTAVNNLGDD114 pKa = 4.57IINNAIGDD122 pKa = 4.01IKK124 pKa = 11.16ASLDD128 pKa = 3.64SLSTCKK134 pKa = 10.5DD135 pKa = 3.35CSIDD139 pKa = 3.76PFPSAIIDD147 pKa = 3.97NIVPTDD153 pKa = 3.59PTTEE157 pKa = 4.45DD158 pKa = 3.44GNGTVSFDD166 pKa = 4.65VYY168 pKa = 11.26LSDD171 pKa = 5.45CNGTSIAVLNPASGTEE187 pKa = 4.32PITLSEE193 pKa = 4.71GINTGLTFTDD203 pKa = 5.1LPPGYY208 pKa = 10.54YY209 pKa = 10.17NIEE212 pKa = 4.32VYY214 pKa = 10.73CDD216 pKa = 3.26TEE218 pKa = 5.84LIDD221 pKa = 3.54NTSFIINDD229 pKa = 3.52YY230 pKa = 11.06DD231 pKa = 3.92SSEE234 pKa = 4.51DD235 pKa = 3.44GGGDD239 pKa = 3.54EE240 pKa = 5.03EE241 pKa = 6.1IIDD244 pKa = 4.46IVTPDD249 pKa = 4.78DD250 pKa = 4.27GSLIPPNSNDD260 pKa = 2.89IPVNDD265 pKa = 4.51GEE267 pKa = 4.66DD268 pKa = 3.64FNHH271 pKa = 6.84ILMITKK277 pKa = 10.4GNLIPEE283 pKa = 4.35NNIQCYY289 pKa = 9.59QNLFIVGSDD298 pKa = 3.46FNLNEE303 pKa = 3.95WTTGCLNSFNPNDD316 pKa = 3.9PQGDD320 pKa = 3.59EE321 pKa = 4.73CYY323 pKa = 10.68VAIKK327 pKa = 9.35RR328 pKa = 11.84TSDD331 pKa = 3.03NTFDD335 pKa = 3.54TTRR338 pKa = 11.84EE339 pKa = 4.16SVSNSVKK346 pKa = 10.02GLNWIKK352 pKa = 10.78AIDD355 pKa = 4.41NITITKK361 pKa = 9.68ILEE364 pKa = 4.24NGAFGYY370 pKa = 10.7GDD372 pKa = 3.48KK373 pKa = 10.83FVLNEE378 pKa = 4.62DD379 pKa = 3.56YY380 pKa = 11.18TNYY383 pKa = 11.11DD384 pKa = 3.94LIISKK389 pKa = 9.54WYY391 pKa = 9.93FNTSAGTAVKK401 pKa = 9.12PQYY404 pKa = 10.29GYY406 pKa = 9.78PIRR409 pKa = 11.84EE410 pKa = 3.91ISDD413 pKa = 4.77LIVTNTWLQKK423 pKa = 8.94QHH425 pKa = 5.66GSNVSYY431 pKa = 10.7SGEE434 pKa = 3.97LTFTDD439 pKa = 4.02TLNGTVTKK447 pKa = 10.61ARR449 pKa = 11.84EE450 pKa = 4.04SDD452 pKa = 3.99FLATMWGVNGIPAPIDD468 pKa = 3.42LNAQVSGTIINMDD481 pKa = 4.28DD482 pKa = 3.55YY483 pKa = 12.02SNADD487 pKa = 3.55FLLINCTYY495 pKa = 11.37ADD497 pKa = 3.47IGNTQLMYY505 pKa = 11.0AKK507 pKa = 10.3DD508 pKa = 3.31IVYY511 pKa = 10.45DD512 pKa = 3.32GTTRR516 pKa = 11.84YY517 pKa = 10.84DD518 pKa = 3.33FVGIGACYY526 pKa = 10.12FSFNVDD532 pKa = 2.88KK533 pKa = 10.53TINIIDD539 pKa = 3.93AALSAEE545 pKa = 4.45IIITSIKK552 pKa = 10.36KK553 pKa = 10.22IILL556 pKa = 3.78
Molecular weight: 60.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8GEA7|W8GEA7_9MOLU Single-stranded DNA-binding protein OS=Candidatus Hepatoplasma crinochetorum Av OX=1427984 GN=ssb PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.38QPNKK9 pKa = 8.29RR10 pKa = 11.84KK11 pKa = 9.61KK12 pKa = 10.05AKK14 pKa = 8.31THH16 pKa = 5.38GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 8.03ATKK25 pKa = 9.69QGQKK29 pKa = 8.5VVKK32 pKa = 9.76SRR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.68GRR39 pKa = 11.84KK40 pKa = 9.07KK41 pKa = 9.85LTNN44 pKa = 3.53
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.38QPNKK9 pKa = 8.29RR10 pKa = 11.84KK11 pKa = 9.61KK12 pKa = 10.05AKK14 pKa = 8.31THH16 pKa = 5.38GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 8.03ATKK25 pKa = 9.69QGQKK29 pKa = 8.5VVKK32 pKa = 9.76SRR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.68GRR39 pKa = 11.84KK40 pKa = 9.07KK41 pKa = 9.85LTNN44 pKa = 3.53
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
201572 |
30 |
1935 |
346.3 |
40.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.943 ± 0.094 | 0.374 ± 0.022 |
5.795 ± 0.102 | 6.576 ± 0.091 |
6.074 ± 0.127 | 4.733 ± 0.114 |
1.106 ± 0.032 | 11.724 ± 0.106 |
11.373 ± 0.214 | 9.678 ± 0.107 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.419 ± 0.037 | 9.137 ± 0.14 |
2.264 ± 0.041 | 3.029 ± 0.059 |
2.531 ± 0.075 | 5.91 ± 0.079 |
4.496 ± 0.115 | 4.118 ± 0.078 |
0.968 ± 0.041 | 4.752 ± 0.096 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
