
Pseudidiomarina planktonica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Idiomarinaceae; Pseudidiomarina
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
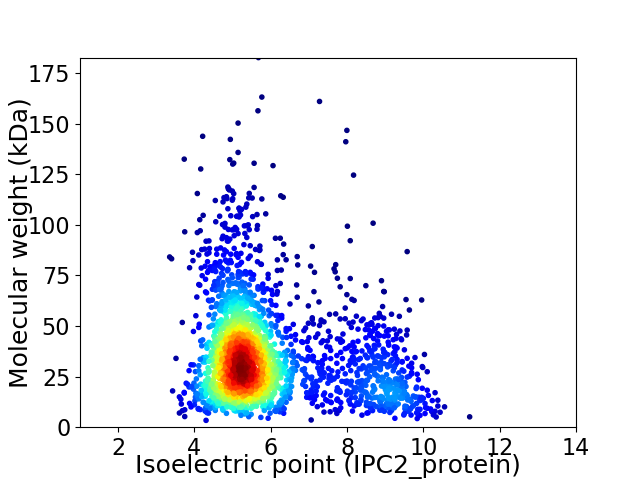
Virtual 2D-PAGE plot for 2385 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
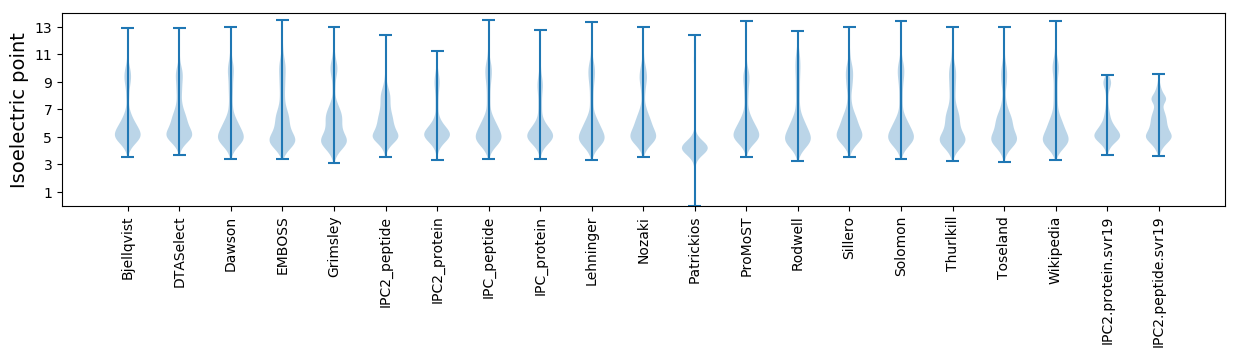
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y6E7Z5|A0A1Y6E7Z5_9GAMM Uncharacterized protein OS=Pseudidiomarina planktonica OX=1323738 GN=SAMN06297229_0167 PE=4 SV=1
MM1 pKa = 7.28YY2 pKa = 10.81SNLFNLSRR10 pKa = 11.84IKK12 pKa = 10.71NLLMLFLVSTVLAACGDD29 pKa = 3.8RR30 pKa = 11.84DD31 pKa = 4.65PILGYY36 pKa = 10.5DD37 pKa = 3.82GNVALSPTVIAVTPADD53 pKa = 3.67NATDD57 pKa = 3.62VAINGASITARR68 pKa = 11.84FSVPVEE74 pKa = 3.83PLSNDD79 pKa = 3.62DD80 pKa = 3.67FTLACSDD87 pKa = 3.78TCTSPSGTVSMNDD100 pKa = 3.08AGTVATFTPVAPAVLDD116 pKa = 3.88GSTLYY121 pKa = 10.19TATVHH126 pKa = 5.96YY127 pKa = 9.92AQSKK131 pKa = 10.4DD132 pKa = 3.31NGLVLEE138 pKa = 4.9NPYY141 pKa = 9.72IWSFTTSVVPEE152 pKa = 4.43TIRR155 pKa = 11.84PRR157 pKa = 11.84VTLTDD162 pKa = 3.7PVTTSSGTTVDD173 pKa = 3.61VPINTSVLAAFNEE186 pKa = 3.97NMAPAMFTEE195 pKa = 4.31ITFTLTCEE203 pKa = 4.54APCMAPQGAVSYY215 pKa = 10.59DD216 pKa = 3.51VSAKK220 pKa = 10.16AAQFKK225 pKa = 9.28PAQDD229 pKa = 4.45LEE231 pKa = 4.36WEE233 pKa = 4.38TTYY236 pKa = 10.6TATISQAVTDD246 pKa = 4.11LAGNEE251 pKa = 4.03LAGNQGPLTEE261 pKa = 4.63ASDD264 pKa = 4.23YY265 pKa = 10.79VWQFTTGSTPDD276 pKa = 3.05ATRR279 pKa = 11.84PRR281 pKa = 11.84VTLTDD286 pKa = 3.84PASTSPANVSVNTSVLAVFNEE307 pKa = 3.66NMAPATFTDD316 pKa = 3.33ATFTLTCDD324 pKa = 3.84APCTGPQGAVSYY336 pKa = 10.64DD337 pKa = 3.51VSAKK341 pKa = 10.16AAQFKK346 pKa = 9.28PAQDD350 pKa = 4.45LEE352 pKa = 4.36WEE354 pKa = 4.38TTYY357 pKa = 10.6TATISQAVTDD367 pKa = 4.11LAGNEE372 pKa = 3.96LAGNQGSVTDD382 pKa = 4.1PSDD385 pKa = 3.72YY386 pKa = 11.17AWEE389 pKa = 4.32FTTGVAPDD397 pKa = 3.56TTRR400 pKa = 11.84PRR402 pKa = 11.84VTLTNPATTSPEE414 pKa = 4.11LEE416 pKa = 3.83TDD418 pKa = 3.67VPVNTSVLAVFNEE431 pKa = 3.61NMVPSTFTDD440 pKa = 3.34TTFTLTCDD448 pKa = 3.91SPCATSLGTVSYY460 pKa = 11.15NVDD463 pKa = 2.93AKK465 pKa = 10.3SARR468 pKa = 11.84FVPAQDD474 pKa = 4.33LEE476 pKa = 5.02AEE478 pKa = 4.38TTYY481 pKa = 10.44TATISRR487 pKa = 11.84SVTDD491 pKa = 3.55LAGNEE496 pKa = 4.03LAGNQGPLTDD506 pKa = 4.2PSDD509 pKa = 4.09YY510 pKa = 10.88VWQFTTGLAPDD521 pKa = 3.77TTRR524 pKa = 11.84PRR526 pKa = 11.84VTLTEE531 pKa = 4.33PEE533 pKa = 4.45TTSSGPTADD542 pKa = 3.59VPVNTSVLAVFNEE555 pKa = 3.63NMAPATLTDD564 pKa = 3.35TTFTLTCDD572 pKa = 3.82SPCAVPLGTVSYY584 pKa = 11.09NVDD587 pKa = 2.93AKK589 pKa = 10.28SARR592 pKa = 11.84FRR594 pKa = 11.84PAQNLEE600 pKa = 3.79AEE602 pKa = 4.5TTYY605 pKa = 10.55TATISQAVTDD615 pKa = 4.11LAGNEE620 pKa = 3.96LAGNQGSVTQPSDD633 pKa = 3.75YY634 pKa = 10.76VWQFTTGLTPDD645 pKa = 4.25TIRR648 pKa = 11.84PRR650 pKa = 11.84VTLTEE655 pKa = 4.37PATSSPGPTANAPVNTSVLAVFNEE679 pKa = 3.63NMVPSMFTDD688 pKa = 3.39TTFTLICEE696 pKa = 4.78APCVAPVGTVSYY708 pKa = 11.22NVDD711 pKa = 3.09TKK713 pKa = 10.45SARR716 pKa = 11.84FSPAQDD722 pKa = 3.97LEE724 pKa = 4.35PEE726 pKa = 4.23TTYY729 pKa = 10.47TATISRR735 pKa = 11.84SVTDD739 pKa = 3.55LAGNEE744 pKa = 4.03LAGNQGPVTDD754 pKa = 4.39PSDD757 pKa = 3.89YY758 pKa = 10.86VWQFTTGLAPDD769 pKa = 3.77TTRR772 pKa = 11.84PRR774 pKa = 11.84VTLTEE779 pKa = 4.23PATTSPGPTADD790 pKa = 3.54VPVNTQIKK798 pKa = 10.1AIFSEE803 pKa = 5.15AISPTTITGTSFTLTCEE820 pKa = 4.56EE821 pKa = 5.09PCTAPIGNVSYY832 pKa = 10.83DD833 pKa = 3.37VSSRR837 pKa = 11.84SAVFTPEE844 pKa = 3.5QNLAEE849 pKa = 4.6GEE851 pKa = 4.62TYY853 pKa = 9.63TATLVSTITDD863 pKa = 3.88LAGNRR868 pKa = 11.84LAGNQGPVTSASDD881 pKa = 4.19YY882 pKa = 9.59IWSFTTTAPIAPTNISVLSTDD903 pKa = 3.97PLDD906 pKa = 4.23AGTMAVCPNASINATFDD923 pKa = 3.19IPSGSRR929 pKa = 11.84LDD931 pKa = 3.74PATVNNMTFLIVEE944 pKa = 4.48DD945 pKa = 4.56ANPLISVLAEE955 pKa = 4.21SIQVDD960 pKa = 3.83VDD962 pKa = 3.31TGTVVTFIPQEE973 pKa = 3.87QLPEE977 pKa = 3.99NVTYY981 pKa = 10.54RR982 pKa = 11.84VTLVGGSDD990 pKa = 3.74GVKK993 pKa = 10.89DD994 pKa = 4.15LMLPGNEE1001 pKa = 4.17MLDD1004 pKa = 3.67DD1005 pKa = 4.1YY1006 pKa = 11.5VWTFTTVAPVEE1017 pKa = 4.48SCLVPVNLQSAAPFGSFGGTAGITNQGVLTVINGDD1052 pKa = 2.94IGTTGTSTIVTGFVSEE1068 pKa = 4.75PGCEE1072 pKa = 3.97YY1073 pKa = 10.72TITTLNEE1080 pKa = 3.68GQVNGKK1086 pKa = 9.22IFTAPPPPTVSCPQDD1101 pKa = 3.13GTAEE1105 pKa = 4.15TEE1107 pKa = 4.58AIATQARR1114 pKa = 11.84LDD1116 pKa = 3.74AEE1118 pKa = 4.18SAYY1121 pKa = 11.25NEE1123 pKa = 3.99LTPANMPGGQNPGNEE1138 pKa = 4.11NLGGLTLAPGIYY1150 pKa = 8.85TAQSGAFRR1158 pKa = 11.84IQGGDD1163 pKa = 3.37LTLDD1167 pKa = 3.51GQGNQNAVWVFQMATTLTVGGPGADD1192 pKa = 3.94FSQSVTLINGAQAKK1206 pKa = 10.2NIFWQVGSAATINAGGGGVMKK1227 pKa = 9.46GTIIAQEE1234 pKa = 4.2GVTISTAGNVDD1245 pKa = 2.7IVTIDD1250 pKa = 3.3GRR1252 pKa = 11.84ALSLGASVTMVNTVINVPADD1272 pKa = 3.18
MM1 pKa = 7.28YY2 pKa = 10.81SNLFNLSRR10 pKa = 11.84IKK12 pKa = 10.71NLLMLFLVSTVLAACGDD29 pKa = 3.8RR30 pKa = 11.84DD31 pKa = 4.65PILGYY36 pKa = 10.5DD37 pKa = 3.82GNVALSPTVIAVTPADD53 pKa = 3.67NATDD57 pKa = 3.62VAINGASITARR68 pKa = 11.84FSVPVEE74 pKa = 3.83PLSNDD79 pKa = 3.62DD80 pKa = 3.67FTLACSDD87 pKa = 3.78TCTSPSGTVSMNDD100 pKa = 3.08AGTVATFTPVAPAVLDD116 pKa = 3.88GSTLYY121 pKa = 10.19TATVHH126 pKa = 5.96YY127 pKa = 9.92AQSKK131 pKa = 10.4DD132 pKa = 3.31NGLVLEE138 pKa = 4.9NPYY141 pKa = 9.72IWSFTTSVVPEE152 pKa = 4.43TIRR155 pKa = 11.84PRR157 pKa = 11.84VTLTDD162 pKa = 3.7PVTTSSGTTVDD173 pKa = 3.61VPINTSVLAAFNEE186 pKa = 3.97NMAPAMFTEE195 pKa = 4.31ITFTLTCEE203 pKa = 4.54APCMAPQGAVSYY215 pKa = 10.59DD216 pKa = 3.51VSAKK220 pKa = 10.16AAQFKK225 pKa = 9.28PAQDD229 pKa = 4.45LEE231 pKa = 4.36WEE233 pKa = 4.38TTYY236 pKa = 10.6TATISQAVTDD246 pKa = 4.11LAGNEE251 pKa = 4.03LAGNQGPLTEE261 pKa = 4.63ASDD264 pKa = 4.23YY265 pKa = 10.79VWQFTTGSTPDD276 pKa = 3.05ATRR279 pKa = 11.84PRR281 pKa = 11.84VTLTDD286 pKa = 3.84PASTSPANVSVNTSVLAVFNEE307 pKa = 3.66NMAPATFTDD316 pKa = 3.33ATFTLTCDD324 pKa = 3.84APCTGPQGAVSYY336 pKa = 10.64DD337 pKa = 3.51VSAKK341 pKa = 10.16AAQFKK346 pKa = 9.28PAQDD350 pKa = 4.45LEE352 pKa = 4.36WEE354 pKa = 4.38TTYY357 pKa = 10.6TATISQAVTDD367 pKa = 4.11LAGNEE372 pKa = 3.96LAGNQGSVTDD382 pKa = 4.1PSDD385 pKa = 3.72YY386 pKa = 11.17AWEE389 pKa = 4.32FTTGVAPDD397 pKa = 3.56TTRR400 pKa = 11.84PRR402 pKa = 11.84VTLTNPATTSPEE414 pKa = 4.11LEE416 pKa = 3.83TDD418 pKa = 3.67VPVNTSVLAVFNEE431 pKa = 3.61NMVPSTFTDD440 pKa = 3.34TTFTLTCDD448 pKa = 3.91SPCATSLGTVSYY460 pKa = 11.15NVDD463 pKa = 2.93AKK465 pKa = 10.3SARR468 pKa = 11.84FVPAQDD474 pKa = 4.33LEE476 pKa = 5.02AEE478 pKa = 4.38TTYY481 pKa = 10.44TATISRR487 pKa = 11.84SVTDD491 pKa = 3.55LAGNEE496 pKa = 4.03LAGNQGPLTDD506 pKa = 4.2PSDD509 pKa = 4.09YY510 pKa = 10.88VWQFTTGLAPDD521 pKa = 3.77TTRR524 pKa = 11.84PRR526 pKa = 11.84VTLTEE531 pKa = 4.33PEE533 pKa = 4.45TTSSGPTADD542 pKa = 3.59VPVNTSVLAVFNEE555 pKa = 3.63NMAPATLTDD564 pKa = 3.35TTFTLTCDD572 pKa = 3.82SPCAVPLGTVSYY584 pKa = 11.09NVDD587 pKa = 2.93AKK589 pKa = 10.28SARR592 pKa = 11.84FRR594 pKa = 11.84PAQNLEE600 pKa = 3.79AEE602 pKa = 4.5TTYY605 pKa = 10.55TATISQAVTDD615 pKa = 4.11LAGNEE620 pKa = 3.96LAGNQGSVTQPSDD633 pKa = 3.75YY634 pKa = 10.76VWQFTTGLTPDD645 pKa = 4.25TIRR648 pKa = 11.84PRR650 pKa = 11.84VTLTEE655 pKa = 4.37PATSSPGPTANAPVNTSVLAVFNEE679 pKa = 3.63NMVPSMFTDD688 pKa = 3.39TTFTLICEE696 pKa = 4.78APCVAPVGTVSYY708 pKa = 11.22NVDD711 pKa = 3.09TKK713 pKa = 10.45SARR716 pKa = 11.84FSPAQDD722 pKa = 3.97LEE724 pKa = 4.35PEE726 pKa = 4.23TTYY729 pKa = 10.47TATISRR735 pKa = 11.84SVTDD739 pKa = 3.55LAGNEE744 pKa = 4.03LAGNQGPVTDD754 pKa = 4.39PSDD757 pKa = 3.89YY758 pKa = 10.86VWQFTTGLAPDD769 pKa = 3.77TTRR772 pKa = 11.84PRR774 pKa = 11.84VTLTEE779 pKa = 4.23PATTSPGPTADD790 pKa = 3.54VPVNTQIKK798 pKa = 10.1AIFSEE803 pKa = 5.15AISPTTITGTSFTLTCEE820 pKa = 4.56EE821 pKa = 5.09PCTAPIGNVSYY832 pKa = 10.83DD833 pKa = 3.37VSSRR837 pKa = 11.84SAVFTPEE844 pKa = 3.5QNLAEE849 pKa = 4.6GEE851 pKa = 4.62TYY853 pKa = 9.63TATLVSTITDD863 pKa = 3.88LAGNRR868 pKa = 11.84LAGNQGPVTSASDD881 pKa = 4.19YY882 pKa = 9.59IWSFTTTAPIAPTNISVLSTDD903 pKa = 3.97PLDD906 pKa = 4.23AGTMAVCPNASINATFDD923 pKa = 3.19IPSGSRR929 pKa = 11.84LDD931 pKa = 3.74PATVNNMTFLIVEE944 pKa = 4.48DD945 pKa = 4.56ANPLISVLAEE955 pKa = 4.21SIQVDD960 pKa = 3.83VDD962 pKa = 3.31TGTVVTFIPQEE973 pKa = 3.87QLPEE977 pKa = 3.99NVTYY981 pKa = 10.54RR982 pKa = 11.84VTLVGGSDD990 pKa = 3.74GVKK993 pKa = 10.89DD994 pKa = 4.15LMLPGNEE1001 pKa = 4.17MLDD1004 pKa = 3.67DD1005 pKa = 4.1YY1006 pKa = 11.5VWTFTTVAPVEE1017 pKa = 4.48SCLVPVNLQSAAPFGSFGGTAGITNQGVLTVINGDD1052 pKa = 2.94IGTTGTSTIVTGFVSEE1068 pKa = 4.75PGCEE1072 pKa = 3.97YY1073 pKa = 10.72TITTLNEE1080 pKa = 3.68GQVNGKK1086 pKa = 9.22IFTAPPPPTVSCPQDD1101 pKa = 3.13GTAEE1105 pKa = 4.15TEE1107 pKa = 4.58AIATQARR1114 pKa = 11.84LDD1116 pKa = 3.74AEE1118 pKa = 4.18SAYY1121 pKa = 11.25NEE1123 pKa = 3.99LTPANMPGGQNPGNEE1138 pKa = 4.11NLGGLTLAPGIYY1150 pKa = 8.85TAQSGAFRR1158 pKa = 11.84IQGGDD1163 pKa = 3.37LTLDD1167 pKa = 3.51GQGNQNAVWVFQMATTLTVGGPGADD1192 pKa = 3.94FSQSVTLINGAQAKK1206 pKa = 10.2NIFWQVGSAATINAGGGGVMKK1227 pKa = 9.46GTIIAQEE1234 pKa = 4.2GVTISTAGNVDD1245 pKa = 2.7IVTIDD1250 pKa = 3.3GRR1252 pKa = 11.84ALSLGASVTMVNTVINVPADD1272 pKa = 3.18
Molecular weight: 132.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y6E641|A0A1Y6E641_9GAMM dGTPase OS=Pseudidiomarina planktonica OX=1323738 GN=SAMN06297229_0057 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
795332 |
32 |
1611 |
333.5 |
36.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.892 ± 0.053 | 0.854 ± 0.015 |
5.67 ± 0.038 | 6.402 ± 0.051 |
3.827 ± 0.03 | 6.945 ± 0.045 |
2.217 ± 0.027 | 5.483 ± 0.039 |
4.15 ± 0.044 | 10.632 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.437 ± 0.024 | 3.766 ± 0.033 |
4.179 ± 0.031 | 5.264 ± 0.05 |
5.491 ± 0.034 | 6.057 ± 0.042 |
5.34 ± 0.034 | 7.209 ± 0.042 |
1.316 ± 0.023 | 2.867 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
