
Pectobacterium phage PP81
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Studiervirinae; Pektosvirus; Pectobacterium virus PP81
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
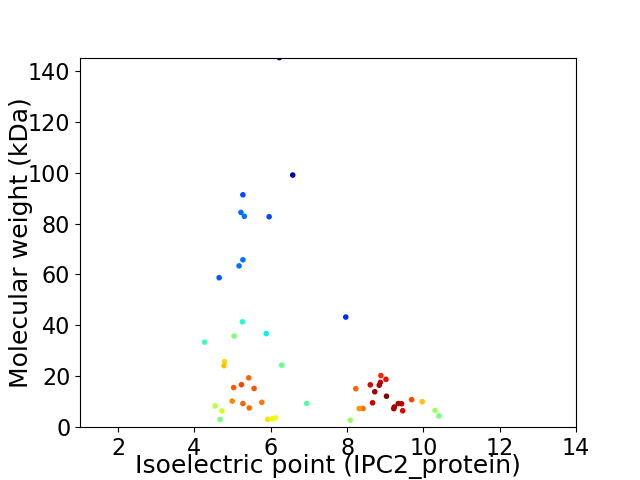
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
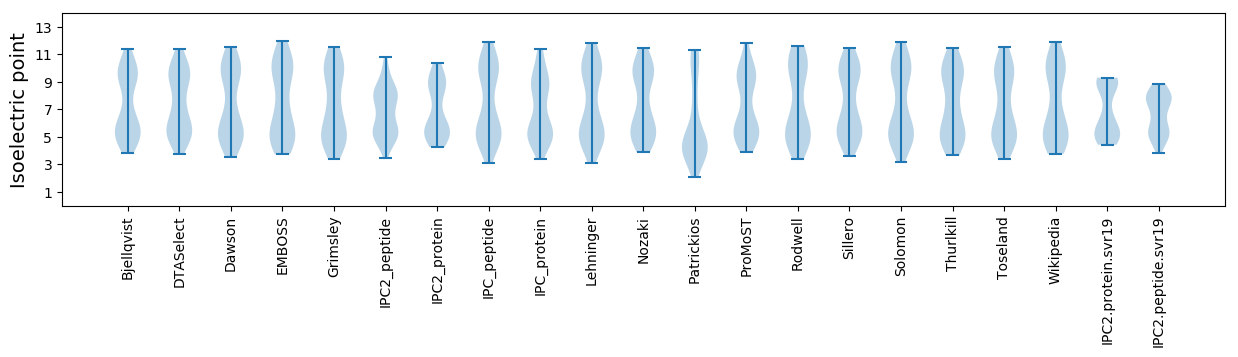
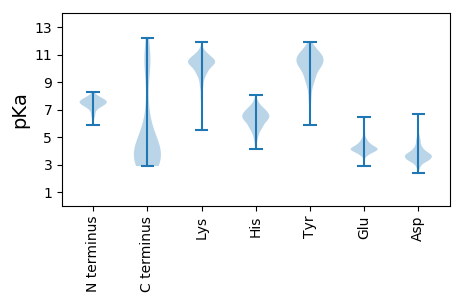
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L7DS89|A0A1L7DS89_9CAUD Holin OS=Pectobacterium phage PP81 OX=1927014 GN=PP81_gp49 PE=3 SV=1
MM1 pKa = 7.7AEE3 pKa = 4.14SNADD7 pKa = 3.26VYY9 pKa = 11.66ASFGVNPAVVTSGSVSEE26 pKa = 5.25HH27 pKa = 4.95EE28 pKa = 4.21QAMLEE33 pKa = 4.18LDD35 pKa = 3.49VSARR39 pKa = 11.84DD40 pKa = 3.39GDD42 pKa = 4.15DD43 pKa = 4.77AITLAEE49 pKa = 4.22MPDD52 pKa = 3.79VQSDD56 pKa = 3.7PNDD59 pKa = 3.84PYY61 pKa = 11.86GNPDD65 pKa = 3.66KK66 pKa = 10.92FADD69 pKa = 4.15PNDD72 pKa = 3.79DD73 pKa = 2.75SRR75 pKa = 11.84MQIRR79 pKa = 11.84INADD83 pKa = 3.23GEE85 pKa = 4.75TVEE88 pKa = 4.86MDD90 pKa = 3.98ADD92 pKa = 3.89GQPIEE97 pKa = 4.89GDD99 pKa = 3.42GDD101 pKa = 3.77DD102 pKa = 4.85TEE104 pKa = 4.46FTPLGEE110 pKa = 4.21PSEE113 pKa = 4.2EE114 pKa = 3.8LSASVEE120 pKa = 3.85LLGQHH125 pKa = 5.62EE126 pKa = 4.86AGFQEE131 pKa = 4.6MVNQAGEE138 pKa = 4.03RR139 pKa = 11.84GLAAEE144 pKa = 4.81SIQRR148 pKa = 11.84IQLEE152 pKa = 4.19YY153 pKa = 9.98QADD156 pKa = 4.75GISEE160 pKa = 3.85QSYY163 pKa = 10.72EE164 pKa = 3.96EE165 pKa = 4.09LAAAGYY171 pKa = 10.38SKK173 pKa = 11.3AFVDD177 pKa = 5.06SYY179 pKa = 11.67IKK181 pKa = 10.65GQEE184 pKa = 4.03ALVDD188 pKa = 4.25GYY190 pKa = 11.11VRR192 pKa = 11.84QVQQMAGGEE201 pKa = 4.18EE202 pKa = 4.49KK203 pKa = 9.79FTNIMSHH210 pKa = 6.84LEE212 pKa = 4.14STNKK216 pKa = 9.32EE217 pKa = 4.03AAEE220 pKa = 4.02SLYY223 pKa = 11.14AALAARR229 pKa = 11.84DD230 pKa = 3.58LATVKK235 pKa = 10.9GILNLAGEE243 pKa = 4.41SRR245 pKa = 11.84AKK247 pKa = 10.72KK248 pKa = 9.73FGKK251 pKa = 8.95PAEE254 pKa = 4.05RR255 pKa = 11.84SVTQRR260 pKa = 11.84ATPSKK265 pKa = 9.59AAVVKK270 pKa = 11.01AEE272 pKa = 4.17GFGSSEE278 pKa = 4.09EE279 pKa = 4.11MVKK282 pKa = 10.77AMSDD286 pKa = 2.97PRR288 pKa = 11.84YY289 pKa = 8.98RR290 pKa = 11.84TDD292 pKa = 2.8SKK294 pKa = 10.77FRR296 pKa = 11.84QEE298 pKa = 4.24VEE300 pKa = 4.05QKK302 pKa = 10.28VAASSFGWW310 pKa = 3.36
MM1 pKa = 7.7AEE3 pKa = 4.14SNADD7 pKa = 3.26VYY9 pKa = 11.66ASFGVNPAVVTSGSVSEE26 pKa = 5.25HH27 pKa = 4.95EE28 pKa = 4.21QAMLEE33 pKa = 4.18LDD35 pKa = 3.49VSARR39 pKa = 11.84DD40 pKa = 3.39GDD42 pKa = 4.15DD43 pKa = 4.77AITLAEE49 pKa = 4.22MPDD52 pKa = 3.79VQSDD56 pKa = 3.7PNDD59 pKa = 3.84PYY61 pKa = 11.86GNPDD65 pKa = 3.66KK66 pKa = 10.92FADD69 pKa = 4.15PNDD72 pKa = 3.79DD73 pKa = 2.75SRR75 pKa = 11.84MQIRR79 pKa = 11.84INADD83 pKa = 3.23GEE85 pKa = 4.75TVEE88 pKa = 4.86MDD90 pKa = 3.98ADD92 pKa = 3.89GQPIEE97 pKa = 4.89GDD99 pKa = 3.42GDD101 pKa = 3.77DD102 pKa = 4.85TEE104 pKa = 4.46FTPLGEE110 pKa = 4.21PSEE113 pKa = 4.2EE114 pKa = 3.8LSASVEE120 pKa = 3.85LLGQHH125 pKa = 5.62EE126 pKa = 4.86AGFQEE131 pKa = 4.6MVNQAGEE138 pKa = 4.03RR139 pKa = 11.84GLAAEE144 pKa = 4.81SIQRR148 pKa = 11.84IQLEE152 pKa = 4.19YY153 pKa = 9.98QADD156 pKa = 4.75GISEE160 pKa = 3.85QSYY163 pKa = 10.72EE164 pKa = 3.96EE165 pKa = 4.09LAAAGYY171 pKa = 10.38SKK173 pKa = 11.3AFVDD177 pKa = 5.06SYY179 pKa = 11.67IKK181 pKa = 10.65GQEE184 pKa = 4.03ALVDD188 pKa = 4.25GYY190 pKa = 11.11VRR192 pKa = 11.84QVQQMAGGEE201 pKa = 4.18EE202 pKa = 4.49KK203 pKa = 9.79FTNIMSHH210 pKa = 6.84LEE212 pKa = 4.14STNKK216 pKa = 9.32EE217 pKa = 4.03AAEE220 pKa = 4.02SLYY223 pKa = 11.14AALAARR229 pKa = 11.84DD230 pKa = 3.58LATVKK235 pKa = 10.9GILNLAGEE243 pKa = 4.41SRR245 pKa = 11.84AKK247 pKa = 10.72KK248 pKa = 9.73FGKK251 pKa = 8.95PAEE254 pKa = 4.05RR255 pKa = 11.84SVTQRR260 pKa = 11.84ATPSKK265 pKa = 9.59AAVVKK270 pKa = 11.01AEE272 pKa = 4.17GFGSSEE278 pKa = 4.09EE279 pKa = 4.11MVKK282 pKa = 10.77AMSDD286 pKa = 2.97PRR288 pKa = 11.84YY289 pKa = 8.98RR290 pKa = 11.84TDD292 pKa = 2.8SKK294 pKa = 10.77FRR296 pKa = 11.84QEE298 pKa = 4.24VEE300 pKa = 4.05QKK302 pKa = 10.28VAASSFGWW310 pKa = 3.36
Molecular weight: 33.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7S8WIF3|A0A7S8WIF3_9CAUD Terminase small subunit OS=Pectobacterium phage PP81 OX=1927014 GN=PP81_gp50 PE=4 SV=1
MM1 pKa = 7.48LKK3 pKa = 10.66APIKK7 pKa = 10.49ASTTIRR13 pKa = 11.84LSDD16 pKa = 4.27TVDD19 pKa = 2.55QWSRR23 pKa = 11.84RR24 pKa = 11.84IHH26 pKa = 5.76VNVRR30 pKa = 11.84NGKK33 pKa = 6.82PTLVYY38 pKa = 9.87RR39 pKa = 11.84WRR41 pKa = 11.84DD42 pKa = 3.35SKK44 pKa = 11.39SPKK47 pKa = 9.74SHH49 pKa = 5.11TQRR52 pKa = 11.84VTLSDD57 pKa = 3.66VQVGRR62 pKa = 11.84LIAALSIATGVAADD76 pKa = 3.92NNEE79 pKa = 3.63SRR81 pKa = 11.84LLVVANGMDD90 pKa = 3.43AQQRR94 pKa = 11.84EE95 pKa = 4.28AGII98 pKa = 4.0
MM1 pKa = 7.48LKK3 pKa = 10.66APIKK7 pKa = 10.49ASTTIRR13 pKa = 11.84LSDD16 pKa = 4.27TVDD19 pKa = 2.55QWSRR23 pKa = 11.84RR24 pKa = 11.84IHH26 pKa = 5.76VNVRR30 pKa = 11.84NGKK33 pKa = 6.82PTLVYY38 pKa = 9.87RR39 pKa = 11.84WRR41 pKa = 11.84DD42 pKa = 3.35SKK44 pKa = 11.39SPKK47 pKa = 9.74SHH49 pKa = 5.11TQRR52 pKa = 11.84VTLSDD57 pKa = 3.66VQVGRR62 pKa = 11.84LIAALSIATGVAADD76 pKa = 3.92NNEE79 pKa = 3.63SRR81 pKa = 11.84LLVVANGMDD90 pKa = 3.43AQQRR94 pKa = 11.84EE95 pKa = 4.28AGII98 pKa = 4.0
Molecular weight: 10.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12669 |
25 |
1338 |
239.0 |
26.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.338 ± 0.54 | 0.924 ± 0.16 |
6.204 ± 0.236 | 6.401 ± 0.264 |
3.923 ± 0.215 | 7.585 ± 0.318 |
1.863 ± 0.197 | 5.004 ± 0.272 |
6.457 ± 0.337 | 8.256 ± 0.29 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.771 ± 0.177 | 4.712 ± 0.266 |
3.584 ± 0.173 | 3.868 ± 0.311 |
5.367 ± 0.233 | 6.086 ± 0.312 |
5.636 ± 0.264 | 7.072 ± 0.246 |
1.405 ± 0.164 | 3.544 ± 0.2 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
