
Nocardiopsis flavescens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; Nocardiopsis
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
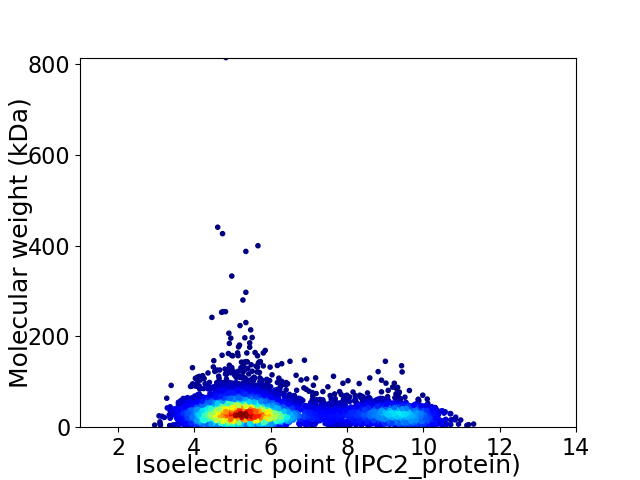
Virtual 2D-PAGE plot for 6204 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6G8V4|A0A1M6G8V4_9ACTN Flp pilus assembly protein CpaB OS=Nocardiopsis flavescens OX=758803 GN=SAMN05421803_103319 PE=4 SV=1
MM1 pKa = 7.15QKK3 pKa = 9.11HH4 pKa = 5.38HH5 pKa = 6.32SRR7 pKa = 11.84RR8 pKa = 11.84TVLGFVAAGALSVPMALSGAVSAGADD34 pKa = 3.69EE35 pKa = 4.94LAPLYY40 pKa = 10.46TSTDD44 pKa = 3.18AVAGEE49 pKa = 4.22WFVVLEE55 pKa = 4.47DD56 pKa = 4.53SISTAAVTPAALGIAAGDD74 pKa = 3.63VNHH77 pKa = 6.98TYY79 pKa = 11.21EE80 pKa = 4.43EE81 pKa = 4.49VFDD84 pKa = 5.12GYY86 pKa = 11.34SATLSTAEE94 pKa = 4.05VQEE97 pKa = 4.7LRR99 pKa = 11.84AQDD102 pKa = 3.16GVAYY106 pKa = 10.07VEE108 pKa = 4.35QVGTAHH114 pKa = 7.04TSVTWGLDD122 pKa = 3.45RR123 pKa = 11.84IDD125 pKa = 5.3QEE127 pKa = 5.42DD128 pKa = 4.22LPLDD132 pKa = 4.04DD133 pKa = 5.66SYY135 pKa = 11.81STTADD140 pKa = 3.2GSGVSAYY147 pKa = 9.8IIDD150 pKa = 3.76TGIDD154 pKa = 3.29PDD156 pKa = 3.96HH157 pKa = 7.48PDD159 pKa = 3.15FEE161 pKa = 5.43GRR163 pKa = 11.84ASSAFDD169 pKa = 3.42AYY171 pKa = 10.67GGGGLDD177 pKa = 3.6VNGHH181 pKa = 4.92GTHH184 pKa = 5.58VAGTIGSATYY194 pKa = 10.31GVAPAADD201 pKa = 3.64LFGVKK206 pKa = 9.7VLSDD210 pKa = 3.99SGSGSYY216 pKa = 11.09DD217 pKa = 3.42DD218 pKa = 5.24VIAGIDD224 pKa = 3.39WVAANAPEE232 pKa = 3.93NSVANLSLGGPASQAVDD249 pKa = 3.56DD250 pKa = 4.95AVNALAGAGVFVAVAAGNEE269 pKa = 4.24GQDD272 pKa = 3.59AGNVSPARR280 pKa = 11.84AEE282 pKa = 4.11GVTTVGASDD291 pKa = 3.37ITDD294 pKa = 3.55TAAYY298 pKa = 9.31FSNHH302 pKa = 4.71GAAVDD307 pKa = 3.93IYY309 pKa = 11.24APGVDD314 pKa = 3.96IEE316 pKa = 4.48STIPGGGVDD325 pKa = 4.74AYY327 pKa = 10.39DD328 pKa = 4.15GTSMASPHH336 pKa = 5.49VAGAAALYY344 pKa = 10.42KK345 pKa = 10.35SANGDD350 pKa = 3.27ADD352 pKa = 3.55QATVQDD358 pKa = 3.68WLVANGSQDD367 pKa = 3.27KK368 pKa = 11.06LSGVPSGTVNTLLNVQGLL386 pKa = 3.81
MM1 pKa = 7.15QKK3 pKa = 9.11HH4 pKa = 5.38HH5 pKa = 6.32SRR7 pKa = 11.84RR8 pKa = 11.84TVLGFVAAGALSVPMALSGAVSAGADD34 pKa = 3.69EE35 pKa = 4.94LAPLYY40 pKa = 10.46TSTDD44 pKa = 3.18AVAGEE49 pKa = 4.22WFVVLEE55 pKa = 4.47DD56 pKa = 4.53SISTAAVTPAALGIAAGDD74 pKa = 3.63VNHH77 pKa = 6.98TYY79 pKa = 11.21EE80 pKa = 4.43EE81 pKa = 4.49VFDD84 pKa = 5.12GYY86 pKa = 11.34SATLSTAEE94 pKa = 4.05VQEE97 pKa = 4.7LRR99 pKa = 11.84AQDD102 pKa = 3.16GVAYY106 pKa = 10.07VEE108 pKa = 4.35QVGTAHH114 pKa = 7.04TSVTWGLDD122 pKa = 3.45RR123 pKa = 11.84IDD125 pKa = 5.3QEE127 pKa = 5.42DD128 pKa = 4.22LPLDD132 pKa = 4.04DD133 pKa = 5.66SYY135 pKa = 11.81STTADD140 pKa = 3.2GSGVSAYY147 pKa = 9.8IIDD150 pKa = 3.76TGIDD154 pKa = 3.29PDD156 pKa = 3.96HH157 pKa = 7.48PDD159 pKa = 3.15FEE161 pKa = 5.43GRR163 pKa = 11.84ASSAFDD169 pKa = 3.42AYY171 pKa = 10.67GGGGLDD177 pKa = 3.6VNGHH181 pKa = 4.92GTHH184 pKa = 5.58VAGTIGSATYY194 pKa = 10.31GVAPAADD201 pKa = 3.64LFGVKK206 pKa = 9.7VLSDD210 pKa = 3.99SGSGSYY216 pKa = 11.09DD217 pKa = 3.42DD218 pKa = 5.24VIAGIDD224 pKa = 3.39WVAANAPEE232 pKa = 3.93NSVANLSLGGPASQAVDD249 pKa = 3.56DD250 pKa = 4.95AVNALAGAGVFVAVAAGNEE269 pKa = 4.24GQDD272 pKa = 3.59AGNVSPARR280 pKa = 11.84AEE282 pKa = 4.11GVTTVGASDD291 pKa = 3.37ITDD294 pKa = 3.55TAAYY298 pKa = 9.31FSNHH302 pKa = 4.71GAAVDD307 pKa = 3.93IYY309 pKa = 11.24APGVDD314 pKa = 3.96IEE316 pKa = 4.48STIPGGGVDD325 pKa = 4.74AYY327 pKa = 10.39DD328 pKa = 4.15GTSMASPHH336 pKa = 5.49VAGAAALYY344 pKa = 10.42KK345 pKa = 10.35SANGDD350 pKa = 3.27ADD352 pKa = 3.55QATVQDD358 pKa = 3.68WLVANGSQDD367 pKa = 3.27KK368 pKa = 11.06LSGVPSGTVNTLLNVQGLL386 pKa = 3.81
Molecular weight: 38.53 kDa
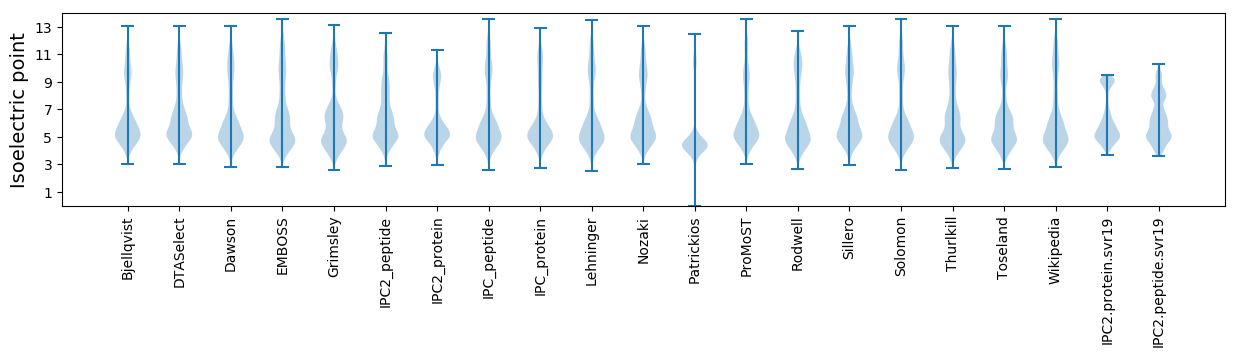
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6QJ49|A0A1M6QJ49_9ACTN Pyruvate dehydrogenase (Quinone) OS=Nocardiopsis flavescens OX=758803 GN=SAMN05421803_11586 PE=3 SV=1
MM1 pKa = 7.65PVNALSALALTAVVLLALSGAATQAKK27 pKa = 9.51KK28 pKa = 10.51ARR30 pKa = 11.84KK31 pKa = 8.67AVAAGRR37 pKa = 11.84RR38 pKa = 11.84TATRR42 pKa = 11.84RR43 pKa = 11.84VRR45 pKa = 11.84AALKK49 pKa = 9.42PRR51 pKa = 11.84TVARR55 pKa = 11.84TRR57 pKa = 11.84RR58 pKa = 11.84KK59 pKa = 7.53TPGRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 3.35
MM1 pKa = 7.65PVNALSALALTAVVLLALSGAATQAKK27 pKa = 9.51KK28 pKa = 10.51ARR30 pKa = 11.84KK31 pKa = 8.67AVAAGRR37 pKa = 11.84RR38 pKa = 11.84TATRR42 pKa = 11.84RR43 pKa = 11.84VRR45 pKa = 11.84AALKK49 pKa = 9.42PRR51 pKa = 11.84TVARR55 pKa = 11.84TRR57 pKa = 11.84RR58 pKa = 11.84KK59 pKa = 7.53TPGRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 3.35
Molecular weight: 6.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2043562 |
28 |
7759 |
329.4 |
35.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.908 ± 0.055 | 0.673 ± 0.009 |
6.195 ± 0.031 | 6.214 ± 0.035 |
2.7 ± 0.017 | 10.16 ± 0.036 |
2.272 ± 0.015 | 2.799 ± 0.024 |
1.312 ± 0.019 | 10.446 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.723 ± 0.015 | 1.546 ± 0.016 |
6.563 ± 0.036 | 2.137 ± 0.017 |
8.71 ± 0.044 | 4.825 ± 0.02 |
5.628 ± 0.031 | 8.82 ± 0.037 |
1.489 ± 0.014 | 1.878 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
