
Macaca mulatta feces associated virus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; unclassified Porprismacovirus
Average proteome isoelectric point is 8.41
Get precalculated fractions of proteins
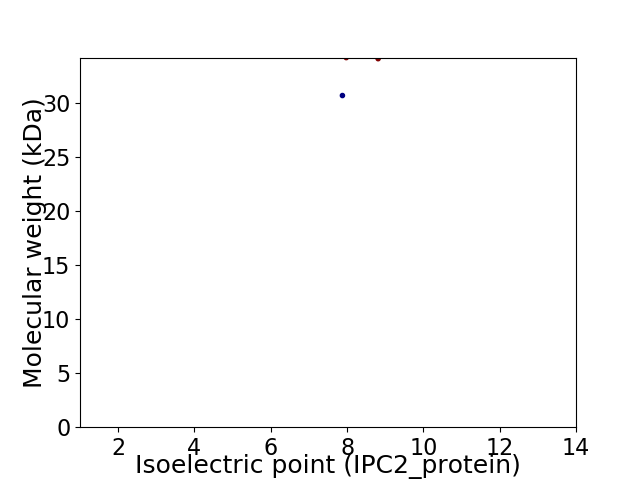
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
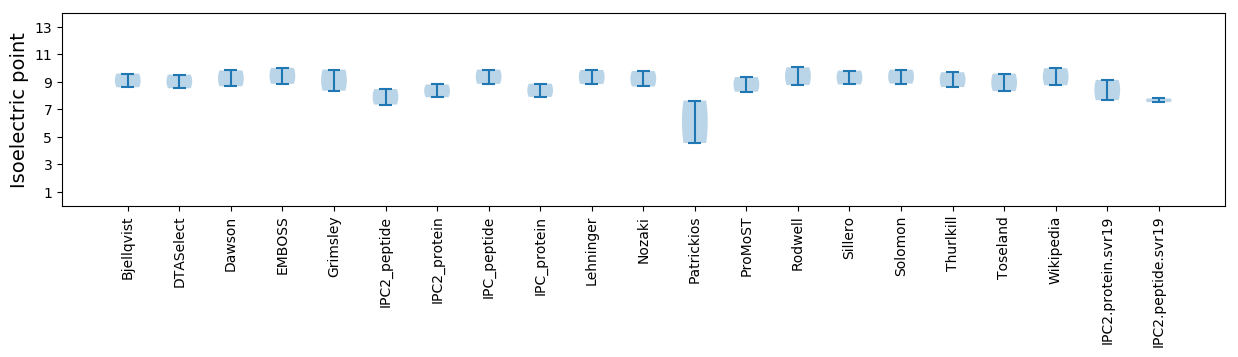
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W5PXG9|A0A1W5PXG9_9VIRU Cap OS=Macaca mulatta feces associated virus 1 OX=2499223 PE=4 SV=1
MM1 pKa = 7.38SKK3 pKa = 9.87IYY5 pKa = 10.13MVTLPRR11 pKa = 11.84TNTYY15 pKa = 11.39AEE17 pKa = 4.48FNALMRR23 pKa = 11.84YY24 pKa = 7.62MRR26 pKa = 11.84QNDD29 pKa = 3.14VHH31 pKa = 7.39KK32 pKa = 9.65WVIGAEE38 pKa = 4.03QGKK41 pKa = 7.78TGYY44 pKa = 9.8DD45 pKa = 2.72HH46 pKa = 6.32WQIRR50 pKa = 11.84LQIGKK55 pKa = 7.13TWEE58 pKa = 3.79EE59 pKa = 4.01VKK61 pKa = 10.62RR62 pKa = 11.84DD63 pKa = 4.19FGAKK67 pKa = 9.58AHH69 pKa = 5.93VEE71 pKa = 4.02EE72 pKa = 6.12ASDD75 pKa = 2.93KK76 pKa = 8.59WTYY79 pKa = 8.12EE80 pKa = 4.36CKK82 pKa = 10.48EE83 pKa = 4.03GMCWKK88 pKa = 10.67SSDD91 pKa = 3.21RR92 pKa = 11.84SEE94 pKa = 4.01NRR96 pKa = 11.84KK97 pKa = 8.73QRR99 pKa = 11.84FGKK102 pKa = 10.02LRR104 pKa = 11.84YY105 pKa = 7.4AQKK108 pKa = 10.58RR109 pKa = 11.84IIEE112 pKa = 4.26HH113 pKa = 7.08LEE115 pKa = 3.9TTNDD119 pKa = 3.35RR120 pKa = 11.84EE121 pKa = 4.38VVVWYY126 pKa = 10.12DD127 pKa = 3.29EE128 pKa = 4.12EE129 pKa = 5.79GNAGKK134 pKa = 10.06SWLTGALWEE143 pKa = 4.68RR144 pKa = 11.84GLAYY148 pKa = 10.03FAVADD153 pKa = 4.1TSGKK157 pKa = 8.4TIVMDD162 pKa = 4.01IASEE166 pKa = 4.03YY167 pKa = 10.46LKK169 pKa = 11.03NGWRR173 pKa = 11.84PYY175 pKa = 10.68VIIDD179 pKa = 3.52IPRR182 pKa = 11.84AGKK185 pKa = 6.86WTPEE189 pKa = 3.79LYY191 pKa = 10.39EE192 pKa = 4.67AIEE195 pKa = 4.45RR196 pKa = 11.84IKK198 pKa = 11.02DD199 pKa = 3.65GLIKK203 pKa = 10.41DD204 pKa = 3.72PRR206 pKa = 11.84YY207 pKa = 10.31SSEE210 pKa = 3.54AVNIHH215 pKa = 5.24GVKK218 pKa = 10.05ILVNTNTMPKK228 pKa = 9.8LDD230 pKa = 4.6KK231 pKa = 10.66LSKK234 pKa = 10.52DD235 pKa = 2.58RR236 pKa = 11.84WKK238 pKa = 10.62IITQTEE244 pKa = 4.31MNNEE248 pKa = 3.92EE249 pKa = 4.41QYY251 pKa = 11.3VDD253 pKa = 3.09TWGGKK258 pKa = 9.71AGAFSS263 pKa = 3.46
MM1 pKa = 7.38SKK3 pKa = 9.87IYY5 pKa = 10.13MVTLPRR11 pKa = 11.84TNTYY15 pKa = 11.39AEE17 pKa = 4.48FNALMRR23 pKa = 11.84YY24 pKa = 7.62MRR26 pKa = 11.84QNDD29 pKa = 3.14VHH31 pKa = 7.39KK32 pKa = 9.65WVIGAEE38 pKa = 4.03QGKK41 pKa = 7.78TGYY44 pKa = 9.8DD45 pKa = 2.72HH46 pKa = 6.32WQIRR50 pKa = 11.84LQIGKK55 pKa = 7.13TWEE58 pKa = 3.79EE59 pKa = 4.01VKK61 pKa = 10.62RR62 pKa = 11.84DD63 pKa = 4.19FGAKK67 pKa = 9.58AHH69 pKa = 5.93VEE71 pKa = 4.02EE72 pKa = 6.12ASDD75 pKa = 2.93KK76 pKa = 8.59WTYY79 pKa = 8.12EE80 pKa = 4.36CKK82 pKa = 10.48EE83 pKa = 4.03GMCWKK88 pKa = 10.67SSDD91 pKa = 3.21RR92 pKa = 11.84SEE94 pKa = 4.01NRR96 pKa = 11.84KK97 pKa = 8.73QRR99 pKa = 11.84FGKK102 pKa = 10.02LRR104 pKa = 11.84YY105 pKa = 7.4AQKK108 pKa = 10.58RR109 pKa = 11.84IIEE112 pKa = 4.26HH113 pKa = 7.08LEE115 pKa = 3.9TTNDD119 pKa = 3.35RR120 pKa = 11.84EE121 pKa = 4.38VVVWYY126 pKa = 10.12DD127 pKa = 3.29EE128 pKa = 4.12EE129 pKa = 5.79GNAGKK134 pKa = 10.06SWLTGALWEE143 pKa = 4.68RR144 pKa = 11.84GLAYY148 pKa = 10.03FAVADD153 pKa = 4.1TSGKK157 pKa = 8.4TIVMDD162 pKa = 4.01IASEE166 pKa = 4.03YY167 pKa = 10.46LKK169 pKa = 11.03NGWRR173 pKa = 11.84PYY175 pKa = 10.68VIIDD179 pKa = 3.52IPRR182 pKa = 11.84AGKK185 pKa = 6.86WTPEE189 pKa = 3.79LYY191 pKa = 10.39EE192 pKa = 4.67AIEE195 pKa = 4.45RR196 pKa = 11.84IKK198 pKa = 11.02DD199 pKa = 3.65GLIKK203 pKa = 10.41DD204 pKa = 3.72PRR206 pKa = 11.84YY207 pKa = 10.31SSEE210 pKa = 3.54AVNIHH215 pKa = 5.24GVKK218 pKa = 10.05ILVNTNTMPKK228 pKa = 9.8LDD230 pKa = 4.6KK231 pKa = 10.66LSKK234 pKa = 10.52DD235 pKa = 2.58RR236 pKa = 11.84WKK238 pKa = 10.62IITQTEE244 pKa = 4.31MNNEE248 pKa = 3.92EE249 pKa = 4.41QYY251 pKa = 11.3VDD253 pKa = 3.09TWGGKK258 pKa = 9.71AGAFSS263 pKa = 3.46
Molecular weight: 30.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W5PXG9|A0A1W5PXG9_9VIRU Cap OS=Macaca mulatta feces associated virus 1 OX=2499223 PE=4 SV=1
MM1 pKa = 8.02IEE3 pKa = 3.96FSRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.55NNHH11 pKa = 4.95GNIAHH16 pKa = 6.4GKK18 pKa = 9.32DD19 pKa = 3.2RR20 pKa = 11.84SITNSGASITIEE32 pKa = 4.1DD33 pKa = 3.67SSITSRR39 pKa = 11.84DD40 pKa = 3.13SRR42 pKa = 11.84ISIPEE47 pKa = 3.9CSVEE51 pKa = 4.24TGHH54 pKa = 5.75SHH56 pKa = 6.53SRR58 pKa = 11.84TTHH61 pKa = 5.45EE62 pKa = 3.94PNINGFSNRR71 pKa = 11.84IPRR74 pKa = 11.84SRR76 pKa = 11.84SEE78 pKa = 3.46ISGIEE83 pKa = 3.95STKK86 pKa = 10.8GSIHH90 pKa = 6.31SRR92 pKa = 11.84GGGHH96 pKa = 6.94RR97 pKa = 11.84EE98 pKa = 3.42NDD100 pKa = 3.92RR101 pKa = 11.84IKK103 pKa = 10.63SLHH106 pKa = 6.15GKK108 pKa = 7.82TSLGHH113 pKa = 6.09SLLPTFSISEE123 pKa = 4.04EE124 pKa = 4.36TIVHH128 pKa = 6.38IEE130 pKa = 4.55LIDD133 pKa = 3.71GNIIIEE139 pKa = 4.31LEE141 pKa = 3.9TRR143 pKa = 11.84IIDD146 pKa = 3.45IYY148 pKa = 11.46DD149 pKa = 3.05RR150 pKa = 11.84TLIHH154 pKa = 6.39NRR156 pKa = 11.84IGNRR160 pKa = 11.84GLPGFEE166 pKa = 4.02PSHH169 pKa = 6.65EE170 pKa = 4.0IRR172 pKa = 11.84HH173 pKa = 5.84GIIGYY178 pKa = 9.55SPVKK182 pKa = 10.62DD183 pKa = 3.18RR184 pKa = 11.84VEE186 pKa = 4.46HH187 pKa = 6.45ILRR190 pKa = 11.84GDD192 pKa = 3.23IPSRR196 pKa = 11.84KK197 pKa = 7.72TDD199 pKa = 3.4LEE201 pKa = 4.54RR202 pKa = 11.84ISRR205 pKa = 11.84KK206 pKa = 9.32HH207 pKa = 6.25RR208 pKa = 11.84STSEE212 pKa = 3.71SNIAIDD218 pKa = 3.77EE219 pKa = 4.21LSILVIHH226 pKa = 7.18RR227 pKa = 11.84KK228 pKa = 6.92TAVHH232 pKa = 6.49LMDD235 pKa = 4.29EE236 pKa = 4.36FRR238 pKa = 11.84LRR240 pKa = 11.84SVDD243 pKa = 2.98TDD245 pKa = 3.37NSHH248 pKa = 7.05FAYY251 pKa = 10.62LSRR254 pKa = 11.84QIISFRR260 pKa = 11.84YY261 pKa = 6.92TDD263 pKa = 3.79KK264 pKa = 11.29NHH266 pKa = 7.19GITFSTCACRR276 pKa = 11.84SDD278 pKa = 4.3RR279 pKa = 11.84RR280 pKa = 11.84ICVRR284 pKa = 11.84RR285 pKa = 11.84TCDD288 pKa = 2.42RR289 pKa = 11.84KK290 pKa = 8.49TFYY293 pKa = 9.78KK294 pKa = 10.41TFFRR298 pKa = 11.84TT299 pKa = 3.37
MM1 pKa = 8.02IEE3 pKa = 3.96FSRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.55NNHH11 pKa = 4.95GNIAHH16 pKa = 6.4GKK18 pKa = 9.32DD19 pKa = 3.2RR20 pKa = 11.84SITNSGASITIEE32 pKa = 4.1DD33 pKa = 3.67SSITSRR39 pKa = 11.84DD40 pKa = 3.13SRR42 pKa = 11.84ISIPEE47 pKa = 3.9CSVEE51 pKa = 4.24TGHH54 pKa = 5.75SHH56 pKa = 6.53SRR58 pKa = 11.84TTHH61 pKa = 5.45EE62 pKa = 3.94PNINGFSNRR71 pKa = 11.84IPRR74 pKa = 11.84SRR76 pKa = 11.84SEE78 pKa = 3.46ISGIEE83 pKa = 3.95STKK86 pKa = 10.8GSIHH90 pKa = 6.31SRR92 pKa = 11.84GGGHH96 pKa = 6.94RR97 pKa = 11.84EE98 pKa = 3.42NDD100 pKa = 3.92RR101 pKa = 11.84IKK103 pKa = 10.63SLHH106 pKa = 6.15GKK108 pKa = 7.82TSLGHH113 pKa = 6.09SLLPTFSISEE123 pKa = 4.04EE124 pKa = 4.36TIVHH128 pKa = 6.38IEE130 pKa = 4.55LIDD133 pKa = 3.71GNIIIEE139 pKa = 4.31LEE141 pKa = 3.9TRR143 pKa = 11.84IIDD146 pKa = 3.45IYY148 pKa = 11.46DD149 pKa = 3.05RR150 pKa = 11.84TLIHH154 pKa = 6.39NRR156 pKa = 11.84IGNRR160 pKa = 11.84GLPGFEE166 pKa = 4.02PSHH169 pKa = 6.65EE170 pKa = 4.0IRR172 pKa = 11.84HH173 pKa = 5.84GIIGYY178 pKa = 9.55SPVKK182 pKa = 10.62DD183 pKa = 3.18RR184 pKa = 11.84VEE186 pKa = 4.46HH187 pKa = 6.45ILRR190 pKa = 11.84GDD192 pKa = 3.23IPSRR196 pKa = 11.84KK197 pKa = 7.72TDD199 pKa = 3.4LEE201 pKa = 4.54RR202 pKa = 11.84ISRR205 pKa = 11.84KK206 pKa = 9.32HH207 pKa = 6.25RR208 pKa = 11.84STSEE212 pKa = 3.71SNIAIDD218 pKa = 3.77EE219 pKa = 4.21LSILVIHH226 pKa = 7.18RR227 pKa = 11.84KK228 pKa = 6.92TAVHH232 pKa = 6.49LMDD235 pKa = 4.29EE236 pKa = 4.36FRR238 pKa = 11.84LRR240 pKa = 11.84SVDD243 pKa = 2.98TDD245 pKa = 3.37NSHH248 pKa = 7.05FAYY251 pKa = 10.62LSRR254 pKa = 11.84QIISFRR260 pKa = 11.84YY261 pKa = 6.92TDD263 pKa = 3.79KK264 pKa = 11.29NHH266 pKa = 7.19GITFSTCACRR276 pKa = 11.84SDD278 pKa = 4.3RR279 pKa = 11.84RR280 pKa = 11.84ICVRR284 pKa = 11.84RR285 pKa = 11.84TCDD288 pKa = 2.42RR289 pKa = 11.84KK290 pKa = 8.49TFYY293 pKa = 9.78KK294 pKa = 10.41TFFRR298 pKa = 11.84TT299 pKa = 3.37
Molecular weight: 34.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
562 |
263 |
299 |
281.0 |
32.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.27 ± 1.643 | 1.246 ± 0.31 |
5.694 ± 0.006 | 7.651 ± 0.698 |
2.847 ± 0.604 | 6.94 ± 0.182 |
4.27 ± 1.512 | 10.32 ± 2.219 |
6.406 ± 1.736 | 5.16 ± 0.104 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.779 ± 0.806 | 4.626 ± 0.041 |
2.491 ± 0.134 | 1.601 ± 0.919 |
9.253 ± 1.78 | 8.897 ± 2.767 |
7.117 ± 0.417 | 4.093 ± 1.028 |
2.135 ± 1.55 | 3.203 ± 1.111 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
