
Capybara microvirus Cap3_SP_410
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.78
Get precalculated fractions of proteins
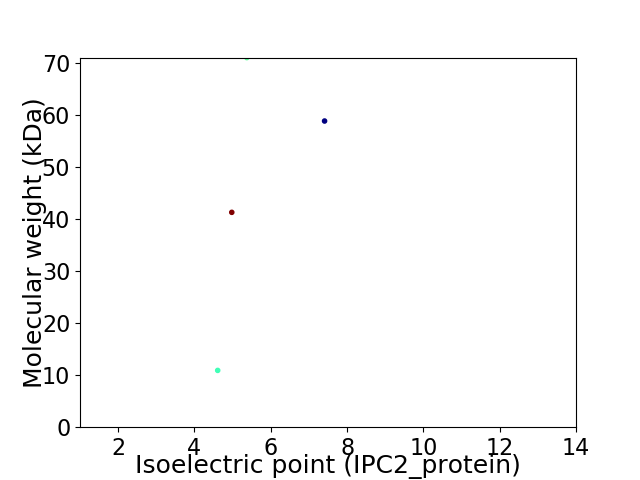
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W4R3|A0A4P8W4R3_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_410 OX=2585446 PE=4 SV=1
MM1 pKa = 7.68AKK3 pKa = 10.58LNIFGFNQDD12 pKa = 3.8AVPLEE17 pKa = 4.46DD18 pKa = 5.21DD19 pKa = 4.28KK20 pKa = 11.8LPHH23 pKa = 7.09RR24 pKa = 11.84ISQVFFDD31 pKa = 4.06EE32 pKa = 4.59EE33 pKa = 3.88QDD35 pKa = 3.28KK36 pKa = 10.86FIPFKK41 pKa = 10.85DD42 pKa = 3.32NPRR45 pKa = 11.84FVSASAVDD53 pKa = 4.39LPSLLAAGIQPGEE66 pKa = 3.88AHH68 pKa = 6.11QEE70 pKa = 3.62ISSLSFVADD79 pKa = 4.55FNRR82 pKa = 11.84SLSSFPQIDD91 pKa = 3.55NQTSNKK97 pKa = 9.59SS98 pKa = 3.19
MM1 pKa = 7.68AKK3 pKa = 10.58LNIFGFNQDD12 pKa = 3.8AVPLEE17 pKa = 4.46DD18 pKa = 5.21DD19 pKa = 4.28KK20 pKa = 11.8LPHH23 pKa = 7.09RR24 pKa = 11.84ISQVFFDD31 pKa = 4.06EE32 pKa = 4.59EE33 pKa = 3.88QDD35 pKa = 3.28KK36 pKa = 10.86FIPFKK41 pKa = 10.85DD42 pKa = 3.32NPRR45 pKa = 11.84FVSASAVDD53 pKa = 4.39LPSLLAAGIQPGEE66 pKa = 3.88AHH68 pKa = 6.11QEE70 pKa = 3.62ISSLSFVADD79 pKa = 4.55FNRR82 pKa = 11.84SLSSFPQIDD91 pKa = 3.55NQTSNKK97 pKa = 9.59SS98 pKa = 3.19
Molecular weight: 10.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4Y2|A0A4P8W4Y2_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_410 OX=2585446 PE=4 SV=1
MM1 pKa = 7.41KK2 pKa = 10.36VLNNSCYY9 pKa = 10.04FSPVNYY15 pKa = 9.64RR16 pKa = 11.84KK17 pKa = 10.56YY18 pKa = 11.11NFLPYY23 pKa = 10.12SSNYY27 pKa = 10.48SYY29 pKa = 11.5DD30 pKa = 3.4LSKK33 pKa = 11.06SLSYY37 pKa = 10.72LVRR40 pKa = 11.84CYY42 pKa = 9.98YY43 pKa = 7.96QWKK46 pKa = 10.41YY47 pKa = 11.03SVEE50 pKa = 4.18LCNGHH55 pKa = 6.65CFLFMLTYY63 pKa = 10.78NDD65 pKa = 3.49NNLPLFDD72 pKa = 5.97DD73 pKa = 5.26FFPCHH78 pKa = 6.19SSKK81 pKa = 10.93DD82 pKa = 3.17IRR84 pKa = 11.84DD85 pKa = 3.42FTKK88 pKa = 10.56RR89 pKa = 11.84LNYY92 pKa = 9.6YY93 pKa = 9.35IKK95 pKa = 10.59SKK97 pKa = 10.46SASFEE102 pKa = 3.5WSYY105 pKa = 11.5FCSKK109 pKa = 10.68EE110 pKa = 3.9FGEE113 pKa = 4.43KK114 pKa = 8.9TKK116 pKa = 10.64RR117 pKa = 11.84PHH119 pKa = 4.79YY120 pKa = 10.43HH121 pKa = 6.44FLLFTDD127 pKa = 5.11FNEE130 pKa = 4.56SVDD133 pKa = 4.17EE134 pKa = 4.06ICRR137 pKa = 11.84LVNLAWITRR146 pKa = 11.84DD147 pKa = 3.14GSKK150 pKa = 10.91GGFVSVSPKK159 pKa = 10.13HH160 pKa = 5.0GAEE163 pKa = 3.71IKK165 pKa = 10.91SFGALKK171 pKa = 10.49YY172 pKa = 9.66VIKK175 pKa = 10.79YY176 pKa = 8.85VIKK179 pKa = 9.66DD180 pKa = 3.29TSTRR184 pKa = 11.84KK185 pKa = 10.36YY186 pKa = 10.5YY187 pKa = 10.85DD188 pKa = 5.04DD189 pKa = 4.6IYY191 pKa = 10.85QHH193 pKa = 7.94IYY195 pKa = 9.36MNNEE199 pKa = 3.36TSFDD203 pKa = 3.48VDD205 pKa = 4.1LDD207 pKa = 3.72MQIYY211 pKa = 9.92YY212 pKa = 10.69YY213 pKa = 11.13NNHH216 pKa = 6.44CNNNNIVDD224 pKa = 3.71NGTIDD229 pKa = 3.9FLCEE233 pKa = 3.38ISKK236 pKa = 11.05SKK238 pKa = 10.57GFQKK242 pKa = 10.47FRR244 pKa = 11.84DD245 pKa = 3.93AYY247 pKa = 9.64ISSKK251 pKa = 9.58TDD253 pKa = 2.61SDD255 pKa = 3.51FRR257 pKa = 11.84QWKK260 pKa = 8.92NEE262 pKa = 4.12CGVKK266 pKa = 9.63PQYY269 pKa = 10.71SKK271 pKa = 11.4RR272 pKa = 11.84LGNFALQHH280 pKa = 6.09ISNNTIKK287 pKa = 10.6IPDD290 pKa = 3.74GEE292 pKa = 4.15NFKK295 pKa = 10.78FVNIPLYY302 pKa = 10.46LYY304 pKa = 10.36RR305 pKa = 11.84KK306 pKa = 9.83LYY308 pKa = 10.51YY309 pKa = 10.64NYY311 pKa = 10.28FQDD314 pKa = 3.81GEE316 pKa = 4.59KK317 pKa = 10.36NVHH320 pKa = 5.74YY321 pKa = 9.75FLNEE325 pKa = 3.55HH326 pKa = 6.04GVALKK331 pKa = 10.29YY332 pKa = 10.62KK333 pKa = 9.88QFNSHH338 pKa = 6.14ICNDD342 pKa = 2.84VCFNYY347 pKa = 10.74DD348 pKa = 4.43LIRR351 pKa = 11.84SSEE354 pKa = 3.82YY355 pKa = 11.38SNILKK360 pKa = 10.59GDD362 pKa = 4.05FSQSYY367 pKa = 10.57DD368 pKa = 3.59FIYY371 pKa = 10.85RR372 pKa = 11.84LTLYY376 pKa = 10.68NSLYY380 pKa = 10.31RR381 pKa = 11.84DD382 pKa = 3.81RR383 pKa = 11.84FCPNLEE389 pKa = 5.11DD390 pKa = 4.02YY391 pKa = 11.41VCDD394 pKa = 3.92IDD396 pKa = 4.68PKK398 pKa = 11.09KK399 pKa = 10.99DD400 pKa = 3.94FIKK403 pKa = 10.88FNTHH407 pKa = 7.32AIFHH411 pKa = 6.27PEE413 pKa = 3.68CHH415 pKa = 6.75DD416 pKa = 3.79FSSLSSSQKK425 pKa = 10.61YY426 pKa = 9.85NLITNFYY433 pKa = 6.92QHH435 pKa = 7.09KK436 pKa = 8.92FSTYY440 pKa = 8.48NNHH443 pKa = 7.02PYY445 pKa = 10.15FEE447 pKa = 4.49EE448 pKa = 4.11VSKK451 pKa = 11.25YY452 pKa = 11.29SNLLYY457 pKa = 10.74DD458 pKa = 4.18LTYY461 pKa = 10.58QYY463 pKa = 11.58YY464 pKa = 10.76FNLFKK469 pKa = 10.76NAQQSYY475 pKa = 9.42DD476 pKa = 3.35FVRR479 pKa = 11.84RR480 pKa = 11.84TRR482 pKa = 11.84QQYY485 pKa = 9.84YY486 pKa = 10.75GEE488 pKa = 4.27TT489 pKa = 3.59
MM1 pKa = 7.41KK2 pKa = 10.36VLNNSCYY9 pKa = 10.04FSPVNYY15 pKa = 9.64RR16 pKa = 11.84KK17 pKa = 10.56YY18 pKa = 11.11NFLPYY23 pKa = 10.12SSNYY27 pKa = 10.48SYY29 pKa = 11.5DD30 pKa = 3.4LSKK33 pKa = 11.06SLSYY37 pKa = 10.72LVRR40 pKa = 11.84CYY42 pKa = 9.98YY43 pKa = 7.96QWKK46 pKa = 10.41YY47 pKa = 11.03SVEE50 pKa = 4.18LCNGHH55 pKa = 6.65CFLFMLTYY63 pKa = 10.78NDD65 pKa = 3.49NNLPLFDD72 pKa = 5.97DD73 pKa = 5.26FFPCHH78 pKa = 6.19SSKK81 pKa = 10.93DD82 pKa = 3.17IRR84 pKa = 11.84DD85 pKa = 3.42FTKK88 pKa = 10.56RR89 pKa = 11.84LNYY92 pKa = 9.6YY93 pKa = 9.35IKK95 pKa = 10.59SKK97 pKa = 10.46SASFEE102 pKa = 3.5WSYY105 pKa = 11.5FCSKK109 pKa = 10.68EE110 pKa = 3.9FGEE113 pKa = 4.43KK114 pKa = 8.9TKK116 pKa = 10.64RR117 pKa = 11.84PHH119 pKa = 4.79YY120 pKa = 10.43HH121 pKa = 6.44FLLFTDD127 pKa = 5.11FNEE130 pKa = 4.56SVDD133 pKa = 4.17EE134 pKa = 4.06ICRR137 pKa = 11.84LVNLAWITRR146 pKa = 11.84DD147 pKa = 3.14GSKK150 pKa = 10.91GGFVSVSPKK159 pKa = 10.13HH160 pKa = 5.0GAEE163 pKa = 3.71IKK165 pKa = 10.91SFGALKK171 pKa = 10.49YY172 pKa = 9.66VIKK175 pKa = 10.79YY176 pKa = 8.85VIKK179 pKa = 9.66DD180 pKa = 3.29TSTRR184 pKa = 11.84KK185 pKa = 10.36YY186 pKa = 10.5YY187 pKa = 10.85DD188 pKa = 5.04DD189 pKa = 4.6IYY191 pKa = 10.85QHH193 pKa = 7.94IYY195 pKa = 9.36MNNEE199 pKa = 3.36TSFDD203 pKa = 3.48VDD205 pKa = 4.1LDD207 pKa = 3.72MQIYY211 pKa = 9.92YY212 pKa = 10.69YY213 pKa = 11.13NNHH216 pKa = 6.44CNNNNIVDD224 pKa = 3.71NGTIDD229 pKa = 3.9FLCEE233 pKa = 3.38ISKK236 pKa = 11.05SKK238 pKa = 10.57GFQKK242 pKa = 10.47FRR244 pKa = 11.84DD245 pKa = 3.93AYY247 pKa = 9.64ISSKK251 pKa = 9.58TDD253 pKa = 2.61SDD255 pKa = 3.51FRR257 pKa = 11.84QWKK260 pKa = 8.92NEE262 pKa = 4.12CGVKK266 pKa = 9.63PQYY269 pKa = 10.71SKK271 pKa = 11.4RR272 pKa = 11.84LGNFALQHH280 pKa = 6.09ISNNTIKK287 pKa = 10.6IPDD290 pKa = 3.74GEE292 pKa = 4.15NFKK295 pKa = 10.78FVNIPLYY302 pKa = 10.46LYY304 pKa = 10.36RR305 pKa = 11.84KK306 pKa = 9.83LYY308 pKa = 10.51YY309 pKa = 10.64NYY311 pKa = 10.28FQDD314 pKa = 3.81GEE316 pKa = 4.59KK317 pKa = 10.36NVHH320 pKa = 5.74YY321 pKa = 9.75FLNEE325 pKa = 3.55HH326 pKa = 6.04GVALKK331 pKa = 10.29YY332 pKa = 10.62KK333 pKa = 9.88QFNSHH338 pKa = 6.14ICNDD342 pKa = 2.84VCFNYY347 pKa = 10.74DD348 pKa = 4.43LIRR351 pKa = 11.84SSEE354 pKa = 3.82YY355 pKa = 11.38SNILKK360 pKa = 10.59GDD362 pKa = 4.05FSQSYY367 pKa = 10.57DD368 pKa = 3.59FIYY371 pKa = 10.85RR372 pKa = 11.84LTLYY376 pKa = 10.68NSLYY380 pKa = 10.31RR381 pKa = 11.84DD382 pKa = 3.81RR383 pKa = 11.84FCPNLEE389 pKa = 5.11DD390 pKa = 4.02YY391 pKa = 11.41VCDD394 pKa = 3.92IDD396 pKa = 4.68PKK398 pKa = 11.09KK399 pKa = 10.99DD400 pKa = 3.94FIKK403 pKa = 10.88FNTHH407 pKa = 7.32AIFHH411 pKa = 6.27PEE413 pKa = 3.68CHH415 pKa = 6.75DD416 pKa = 3.79FSSLSSSQKK425 pKa = 10.61YY426 pKa = 9.85NLITNFYY433 pKa = 6.92QHH435 pKa = 7.09KK436 pKa = 8.92FSTYY440 pKa = 8.48NNHH443 pKa = 7.02PYY445 pKa = 10.15FEE447 pKa = 4.49EE448 pKa = 4.11VSKK451 pKa = 11.25YY452 pKa = 11.29SNLLYY457 pKa = 10.74DD458 pKa = 4.18LTYY461 pKa = 10.58QYY463 pKa = 11.58YY464 pKa = 10.76FNLFKK469 pKa = 10.76NAQQSYY475 pKa = 9.42DD476 pKa = 3.35FVRR479 pKa = 11.84RR480 pKa = 11.84TRR482 pKa = 11.84QQYY485 pKa = 9.84YY486 pKa = 10.75GEE488 pKa = 4.27TT489 pKa = 3.59
Molecular weight: 58.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1575 |
98 |
617 |
393.8 |
45.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.46 ± 2.056 | 1.778 ± 0.649 |
6.476 ± 0.501 | 4.762 ± 1.016 |
6.349 ± 1.199 | 4.635 ± 0.668 |
2.286 ± 0.47 | 5.397 ± 0.602 |
5.714 ± 0.912 | 8.19 ± 0.342 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.587 ± 0.342 | 8.127 ± 1.161 |
3.302 ± 0.753 | 5.524 ± 0.81 |
3.937 ± 0.09 | 9.016 ± 0.778 |
4.063 ± 0.696 | 5.397 ± 0.741 |
1.079 ± 0.182 | 6.921 ± 1.873 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
