
Natronoarchaeum philippinense
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Natronoarchaeum
Average proteome isoelectric point is 4.79
Get precalculated fractions of proteins
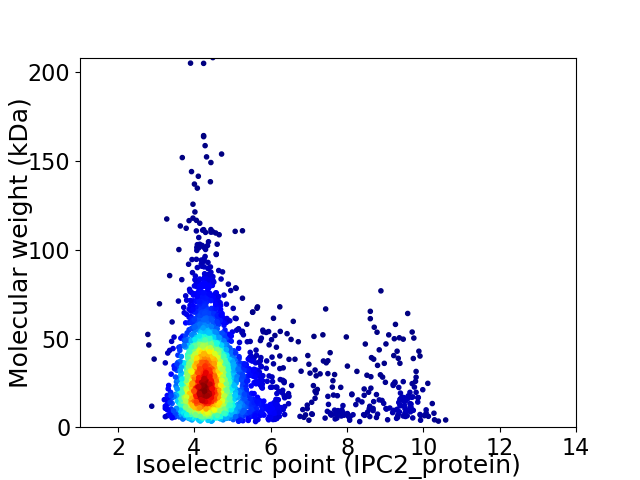
Virtual 2D-PAGE plot for 3284 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A285NWS5|A0A285NWS5_9EURY Uncharacterized protein OS=Natronoarchaeum philippinense OX=558529 GN=SAMN06269185_1491 PE=4 SV=1
MM1 pKa = 5.91TTRR4 pKa = 11.84WAEE7 pKa = 3.35WDD9 pKa = 4.22HH10 pKa = 6.22IVKK13 pKa = 9.88IDD15 pKa = 3.66PDD17 pKa = 3.47KK18 pKa = 11.11TLVEE22 pKa = 4.48GEE24 pKa = 4.34TFEE27 pKa = 5.39DD28 pKa = 3.95VCEE31 pKa = 4.24TGTDD35 pKa = 3.65AIEE38 pKa = 4.22IGGTTGMTTDD48 pKa = 2.83KK49 pKa = 9.06MQRR52 pKa = 11.84VIDD55 pKa = 3.67ACATHH60 pKa = 7.15DD61 pKa = 3.69VPLYY65 pKa = 10.3LEE67 pKa = 4.45PSAPEE72 pKa = 3.63VAVNDD77 pKa = 3.99EE78 pKa = 4.45ALDD81 pKa = 4.55GYY83 pKa = 10.5LIPTVLNAGDD93 pKa = 4.08PFWTVGAHH101 pKa = 6.2KK102 pKa = 9.69EE103 pKa = 3.88WARR106 pKa = 11.84MEE108 pKa = 6.03DD109 pKa = 3.82DD110 pKa = 5.06LPWDD114 pKa = 4.93RR115 pKa = 11.84ITTEE119 pKa = 4.51AYY121 pKa = 10.49VVMNPEE127 pKa = 4.11ASVATYY133 pKa = 10.19TEE135 pKa = 4.47ADD137 pKa = 4.04CDD139 pKa = 4.16LSPDD143 pKa = 3.57DD144 pKa = 3.54VAAYY148 pKa = 10.56AEE150 pKa = 4.47VAEE153 pKa = 4.4EE154 pKa = 4.08MFGQEE159 pKa = 3.39IVYY162 pKa = 9.94IEE164 pKa = 4.02YY165 pKa = 10.61SGTLGDD171 pKa = 3.82PAVVEE176 pKa = 4.38AAADD180 pKa = 3.81ATDD183 pKa = 4.3DD184 pKa = 3.6AALFYY189 pKa = 11.19GGGVHH194 pKa = 7.6DD195 pKa = 5.24YY196 pKa = 11.41DD197 pKa = 4.4SAHH200 pKa = 6.21QMGEE204 pKa = 3.87HH205 pKa = 6.79ADD207 pKa = 3.74VVVVGDD213 pKa = 3.91LVHH216 pKa = 7.21AEE218 pKa = 3.96GVDD221 pKa = 3.53AVEE224 pKa = 4.14EE225 pKa = 4.33TVRR228 pKa = 11.84GAKK231 pKa = 9.94DD232 pKa = 2.87AA233 pKa = 4.34
MM1 pKa = 5.91TTRR4 pKa = 11.84WAEE7 pKa = 3.35WDD9 pKa = 4.22HH10 pKa = 6.22IVKK13 pKa = 9.88IDD15 pKa = 3.66PDD17 pKa = 3.47KK18 pKa = 11.11TLVEE22 pKa = 4.48GEE24 pKa = 4.34TFEE27 pKa = 5.39DD28 pKa = 3.95VCEE31 pKa = 4.24TGTDD35 pKa = 3.65AIEE38 pKa = 4.22IGGTTGMTTDD48 pKa = 2.83KK49 pKa = 9.06MQRR52 pKa = 11.84VIDD55 pKa = 3.67ACATHH60 pKa = 7.15DD61 pKa = 3.69VPLYY65 pKa = 10.3LEE67 pKa = 4.45PSAPEE72 pKa = 3.63VAVNDD77 pKa = 3.99EE78 pKa = 4.45ALDD81 pKa = 4.55GYY83 pKa = 10.5LIPTVLNAGDD93 pKa = 4.08PFWTVGAHH101 pKa = 6.2KK102 pKa = 9.69EE103 pKa = 3.88WARR106 pKa = 11.84MEE108 pKa = 6.03DD109 pKa = 3.82DD110 pKa = 5.06LPWDD114 pKa = 4.93RR115 pKa = 11.84ITTEE119 pKa = 4.51AYY121 pKa = 10.49VVMNPEE127 pKa = 4.11ASVATYY133 pKa = 10.19TEE135 pKa = 4.47ADD137 pKa = 4.04CDD139 pKa = 4.16LSPDD143 pKa = 3.57DD144 pKa = 3.54VAAYY148 pKa = 10.56AEE150 pKa = 4.47VAEE153 pKa = 4.4EE154 pKa = 4.08MFGQEE159 pKa = 3.39IVYY162 pKa = 9.94IEE164 pKa = 4.02YY165 pKa = 10.61SGTLGDD171 pKa = 3.82PAVVEE176 pKa = 4.38AAADD180 pKa = 3.81ATDD183 pKa = 4.3DD184 pKa = 3.6AALFYY189 pKa = 11.19GGGVHH194 pKa = 7.6DD195 pKa = 5.24YY196 pKa = 11.41DD197 pKa = 4.4SAHH200 pKa = 6.21QMGEE204 pKa = 3.87HH205 pKa = 6.79ADD207 pKa = 3.74VVVVGDD213 pKa = 3.91LVHH216 pKa = 7.21AEE218 pKa = 3.96GVDD221 pKa = 3.53AVEE224 pKa = 4.14EE225 pKa = 4.33TVRR228 pKa = 11.84GAKK231 pKa = 9.94DD232 pKa = 2.87AA233 pKa = 4.34
Molecular weight: 25.16 kDa
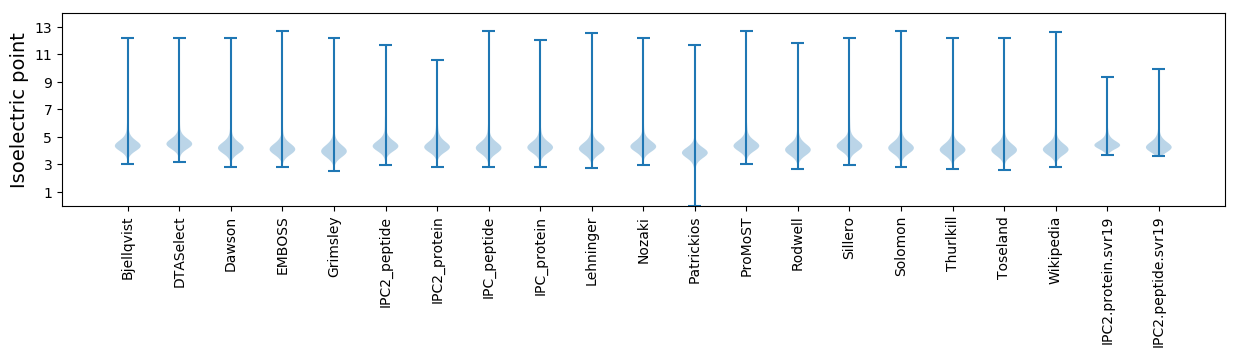
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A285N1W4|A0A285N1W4_9EURY C2H2-type domain-containing protein OS=Natronoarchaeum philippinense OX=558529 GN=SAMN06269185_0274 PE=4 SV=1
MM1 pKa = 7.45NPTIGVDD8 pKa = 3.34ALSGAEE14 pKa = 4.03GVLGYY19 pKa = 11.44ALVFVLAAIPVVEE32 pKa = 4.19VLVVVPAGVAVGLNPPLVALAAFAGNLATVLLVLAGSDD70 pKa = 2.95RR71 pKa = 11.84ALAFVRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84FGSADD84 pKa = 3.41DD85 pKa = 4.86DD86 pKa = 4.15EE87 pKa = 4.78PSKK90 pKa = 10.56RR91 pKa = 11.84VRR93 pKa = 11.84RR94 pKa = 11.84AKK96 pKa = 10.38RR97 pKa = 11.84LWRR100 pKa = 11.84QYY102 pKa = 8.39GTAGLALAAPITTGAHH118 pKa = 5.98LAAVLALSLGSKK130 pKa = 9.57RR131 pKa = 11.84RR132 pKa = 11.84AVATWMATSLLAWTAALATVSVYY155 pKa = 10.73GVDD158 pKa = 3.38SLRR161 pKa = 11.84GLFF164 pKa = 3.95
MM1 pKa = 7.45NPTIGVDD8 pKa = 3.34ALSGAEE14 pKa = 4.03GVLGYY19 pKa = 11.44ALVFVLAAIPVVEE32 pKa = 4.19VLVVVPAGVAVGLNPPLVALAAFAGNLATVLLVLAGSDD70 pKa = 2.95RR71 pKa = 11.84ALAFVRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84FGSADD84 pKa = 3.41DD85 pKa = 4.86DD86 pKa = 4.15EE87 pKa = 4.78PSKK90 pKa = 10.56RR91 pKa = 11.84VRR93 pKa = 11.84RR94 pKa = 11.84AKK96 pKa = 10.38RR97 pKa = 11.84LWRR100 pKa = 11.84QYY102 pKa = 8.39GTAGLALAAPITTGAHH118 pKa = 5.98LAAVLALSLGSKK130 pKa = 9.57RR131 pKa = 11.84RR132 pKa = 11.84AVATWMATSLLAWTAALATVSVYY155 pKa = 10.73GVDD158 pKa = 3.38SLRR161 pKa = 11.84GLFF164 pKa = 3.95
Molecular weight: 16.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
928822 |
27 |
1889 |
282.8 |
30.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.236 ± 0.072 | 0.694 ± 0.014 |
8.981 ± 0.057 | 8.701 ± 0.061 |
3.201 ± 0.027 | 8.508 ± 0.044 |
1.975 ± 0.022 | 4.126 ± 0.03 |
1.799 ± 0.028 | 8.791 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.758 ± 0.018 | 2.292 ± 0.029 |
4.541 ± 0.026 | 2.695 ± 0.029 |
6.477 ± 0.043 | 5.543 ± 0.038 |
6.025 ± 0.033 | 8.838 ± 0.038 |
1.143 ± 0.018 | 2.676 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
