
Sclerotium hydrophilum virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Curvulaviridae; Orthocurvulavirus; Sclerotium hydrophilum orthocurvulavirus 1
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
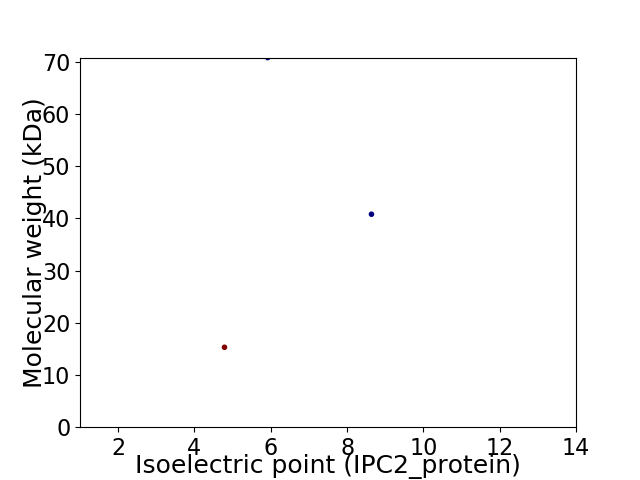
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
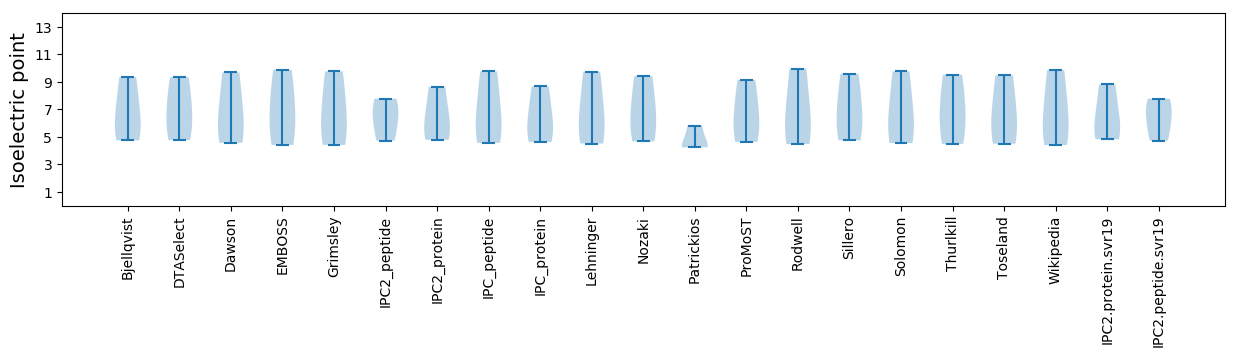
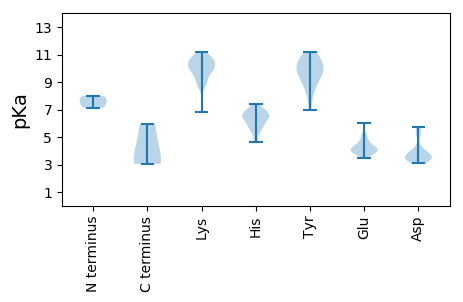
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B3SH34|A0A1B3SH34_9VIRU Putative RNA-dependent RNA polymerase OS=Sclerotium hydrophilum virus 1 OX=1895000 PE=4 SV=1
MM1 pKa = 7.14GWANGRR7 pKa = 11.84RR8 pKa = 11.84SPQDD12 pKa = 3.26VTWACFLYY20 pKa = 9.56YY21 pKa = 9.15WSRR24 pKa = 11.84MSAPPPLDD32 pKa = 3.86LLILEE37 pKa = 4.38EE38 pKa = 4.48HH39 pKa = 6.55AGEE42 pKa = 4.11PQVVISPEE50 pKa = 3.93APLVWRR56 pKa = 11.84MVAAWILRR64 pKa = 11.84VVGGDD69 pKa = 3.4VEE71 pKa = 4.18QLRR74 pKa = 11.84NLVGIVRR81 pKa = 11.84RR82 pKa = 11.84GDD84 pKa = 3.09MLTWIRR90 pKa = 11.84RR91 pKa = 11.84KK92 pKa = 10.24DD93 pKa = 3.23GRR95 pKa = 11.84AFFGNPSQPQAEE107 pKa = 4.25ALSRR111 pKa = 11.84LPEE114 pKa = 4.24PQWVEE119 pKa = 3.59ATEE122 pKa = 4.09EE123 pKa = 4.28EE124 pKa = 4.74EE125 pKa = 4.19GVEE128 pKa = 4.0MQEE131 pKa = 4.84GGTDD135 pKa = 3.04
MM1 pKa = 7.14GWANGRR7 pKa = 11.84RR8 pKa = 11.84SPQDD12 pKa = 3.26VTWACFLYY20 pKa = 9.56YY21 pKa = 9.15WSRR24 pKa = 11.84MSAPPPLDD32 pKa = 3.86LLILEE37 pKa = 4.38EE38 pKa = 4.48HH39 pKa = 6.55AGEE42 pKa = 4.11PQVVISPEE50 pKa = 3.93APLVWRR56 pKa = 11.84MVAAWILRR64 pKa = 11.84VVGGDD69 pKa = 3.4VEE71 pKa = 4.18QLRR74 pKa = 11.84NLVGIVRR81 pKa = 11.84RR82 pKa = 11.84GDD84 pKa = 3.09MLTWIRR90 pKa = 11.84RR91 pKa = 11.84KK92 pKa = 10.24DD93 pKa = 3.23GRR95 pKa = 11.84AFFGNPSQPQAEE107 pKa = 4.25ALSRR111 pKa = 11.84LPEE114 pKa = 4.24PQWVEE119 pKa = 3.59ATEE122 pKa = 4.09EE123 pKa = 4.28EE124 pKa = 4.74EE125 pKa = 4.19GVEE128 pKa = 4.0MQEE131 pKa = 4.84GGTDD135 pKa = 3.04
Molecular weight: 15.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B3SH26|A0A1B3SH26_9VIRU Uncharacterized protein OS=Sclerotium hydrophilum virus 1 OX=1895000 PE=4 SV=1
MM1 pKa = 7.99ADD3 pKa = 3.49FQDD6 pKa = 3.17IPAARR11 pKa = 11.84RR12 pKa = 11.84AQGAGARR19 pKa = 11.84PTMAARR25 pKa = 11.84FGGAGGPVDD34 pKa = 3.83YY35 pKa = 10.24TDD37 pKa = 5.32RR38 pKa = 11.84PAHH41 pKa = 6.96DD42 pKa = 5.59DD43 pKa = 4.17EE44 pKa = 5.18ILPLPSPDD52 pKa = 4.04DD53 pKa = 3.72SFEE56 pKa = 4.17VLVDD60 pKa = 3.39KK61 pKa = 11.19LEE63 pKa = 4.31RR64 pKa = 11.84LAKK67 pKa = 9.91VVRR70 pKa = 11.84DD71 pKa = 3.93NKK73 pKa = 10.85SLALYY78 pKa = 9.19TGFHH82 pKa = 6.29ATPGGAINPVDD93 pKa = 3.47VAKK96 pKa = 10.7RR97 pKa = 11.84QFTTKK102 pKa = 9.91EE103 pKa = 3.62RR104 pKa = 11.84EE105 pKa = 4.08QYY107 pKa = 11.16AEE109 pKa = 3.96WEE111 pKa = 4.53AGSEE115 pKa = 4.34LPPFDD120 pKa = 3.86WNRR123 pKa = 11.84DD124 pKa = 3.59RR125 pKa = 11.84LPVAGKK131 pKa = 10.02DD132 pKa = 3.7GNPATLSKK140 pKa = 9.85WAEE143 pKa = 3.92ALSILWDD150 pKa = 3.54TKK152 pKa = 11.04GADD155 pKa = 3.34NGNVVWFKK163 pKa = 10.85KK164 pKa = 9.02SHH166 pKa = 7.01PYY168 pKa = 10.54LIPLVTAMVGVINARR183 pKa = 11.84KK184 pKa = 9.79GLTSGQAALSEE195 pKa = 4.35MEE197 pKa = 3.92MAEE200 pKa = 4.03YY201 pKa = 8.74QTARR205 pKa = 11.84KK206 pKa = 8.7IAAEE210 pKa = 3.88ATDD213 pKa = 4.02NIRR216 pKa = 11.84ITMEE220 pKa = 3.44AMRR223 pKa = 11.84RR224 pKa = 11.84PTEE227 pKa = 3.9RR228 pKa = 11.84MSAHH232 pKa = 5.69IQVIQEE238 pKa = 4.27RR239 pKa = 11.84SRR241 pKa = 11.84QLGNKK246 pKa = 9.35IPGKK250 pKa = 10.32DD251 pKa = 3.17RR252 pKa = 11.84GDD254 pKa = 3.44GPFNKK259 pKa = 9.43EE260 pKa = 3.5RR261 pKa = 11.84IALGKK266 pKa = 10.13RR267 pKa = 11.84RR268 pKa = 11.84VGVDD272 pKa = 3.6FEE274 pKa = 4.58VTHH277 pKa = 5.94NRR279 pKa = 11.84GAGRR283 pKa = 11.84EE284 pKa = 3.82VRR286 pKa = 11.84TRR288 pKa = 11.84HH289 pKa = 7.0DD290 pKa = 3.34IQQALAMPQRR300 pKa = 11.84PVASTSTTAPRR311 pKa = 11.84APPAPPTGSGSGTGAPGGFARR332 pKa = 11.84WGFGGSTGGFGSSLSQPAPPEE353 pKa = 4.23APPAGAQSPTYY364 pKa = 9.98RR365 pKa = 11.84PASPPPPSDD374 pKa = 3.37VPMAPGHH381 pKa = 5.91
MM1 pKa = 7.99ADD3 pKa = 3.49FQDD6 pKa = 3.17IPAARR11 pKa = 11.84RR12 pKa = 11.84AQGAGARR19 pKa = 11.84PTMAARR25 pKa = 11.84FGGAGGPVDD34 pKa = 3.83YY35 pKa = 10.24TDD37 pKa = 5.32RR38 pKa = 11.84PAHH41 pKa = 6.96DD42 pKa = 5.59DD43 pKa = 4.17EE44 pKa = 5.18ILPLPSPDD52 pKa = 4.04DD53 pKa = 3.72SFEE56 pKa = 4.17VLVDD60 pKa = 3.39KK61 pKa = 11.19LEE63 pKa = 4.31RR64 pKa = 11.84LAKK67 pKa = 9.91VVRR70 pKa = 11.84DD71 pKa = 3.93NKK73 pKa = 10.85SLALYY78 pKa = 9.19TGFHH82 pKa = 6.29ATPGGAINPVDD93 pKa = 3.47VAKK96 pKa = 10.7RR97 pKa = 11.84QFTTKK102 pKa = 9.91EE103 pKa = 3.62RR104 pKa = 11.84EE105 pKa = 4.08QYY107 pKa = 11.16AEE109 pKa = 3.96WEE111 pKa = 4.53AGSEE115 pKa = 4.34LPPFDD120 pKa = 3.86WNRR123 pKa = 11.84DD124 pKa = 3.59RR125 pKa = 11.84LPVAGKK131 pKa = 10.02DD132 pKa = 3.7GNPATLSKK140 pKa = 9.85WAEE143 pKa = 3.92ALSILWDD150 pKa = 3.54TKK152 pKa = 11.04GADD155 pKa = 3.34NGNVVWFKK163 pKa = 10.85KK164 pKa = 9.02SHH166 pKa = 7.01PYY168 pKa = 10.54LIPLVTAMVGVINARR183 pKa = 11.84KK184 pKa = 9.79GLTSGQAALSEE195 pKa = 4.35MEE197 pKa = 3.92MAEE200 pKa = 4.03YY201 pKa = 8.74QTARR205 pKa = 11.84KK206 pKa = 8.7IAAEE210 pKa = 3.88ATDD213 pKa = 4.02NIRR216 pKa = 11.84ITMEE220 pKa = 3.44AMRR223 pKa = 11.84RR224 pKa = 11.84PTEE227 pKa = 3.9RR228 pKa = 11.84MSAHH232 pKa = 5.69IQVIQEE238 pKa = 4.27RR239 pKa = 11.84SRR241 pKa = 11.84QLGNKK246 pKa = 9.35IPGKK250 pKa = 10.32DD251 pKa = 3.17RR252 pKa = 11.84GDD254 pKa = 3.44GPFNKK259 pKa = 9.43EE260 pKa = 3.5RR261 pKa = 11.84IALGKK266 pKa = 10.13RR267 pKa = 11.84RR268 pKa = 11.84VGVDD272 pKa = 3.6FEE274 pKa = 4.58VTHH277 pKa = 5.94NRR279 pKa = 11.84GAGRR283 pKa = 11.84EE284 pKa = 3.82VRR286 pKa = 11.84TRR288 pKa = 11.84HH289 pKa = 7.0DD290 pKa = 3.34IQQALAMPQRR300 pKa = 11.84PVASTSTTAPRR311 pKa = 11.84APPAPPTGSGSGTGAPGGFARR332 pKa = 11.84WGFGGSTGGFGSSLSQPAPPEE353 pKa = 4.23APPAGAQSPTYY364 pKa = 9.98RR365 pKa = 11.84PASPPPPSDD374 pKa = 3.37VPMAPGHH381 pKa = 5.91
Molecular weight: 40.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1140 |
135 |
624 |
380.0 |
42.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.298 ± 1.705 | 1.14 ± 0.595 |
5.965 ± 0.438 | 7.193 ± 1.117 |
4.123 ± 0.775 | 8.684 ± 0.778 |
2.018 ± 0.372 | 4.035 ± 0.216 |
4.737 ± 1.158 | 7.544 ± 1.099 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.07 ± 0.243 | 3.158 ± 0.31 |
6.93 ± 1.732 | 3.246 ± 0.603 |
7.719 ± 0.412 | 5.351 ± 0.279 |
4.912 ± 0.663 | 6.316 ± 0.759 |
1.754 ± 0.899 | 2.807 ± 0.771 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
