
Cucumber mosaic virus (strain Q) (CMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Cucumovirus; Cucumber mosaic virus
Average proteome isoelectric point is 7.82
Get precalculated fractions of proteins
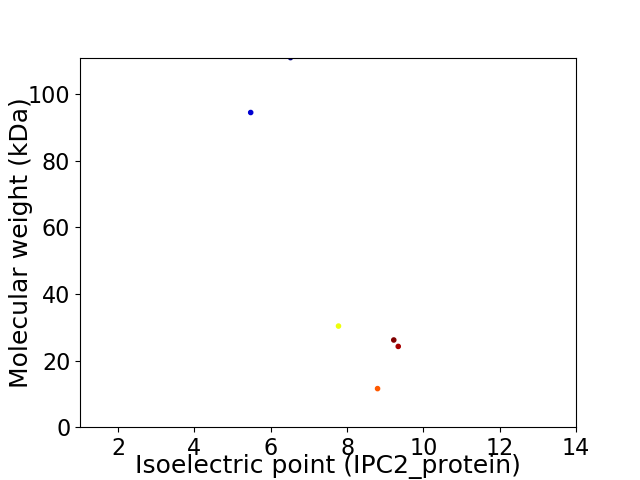
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
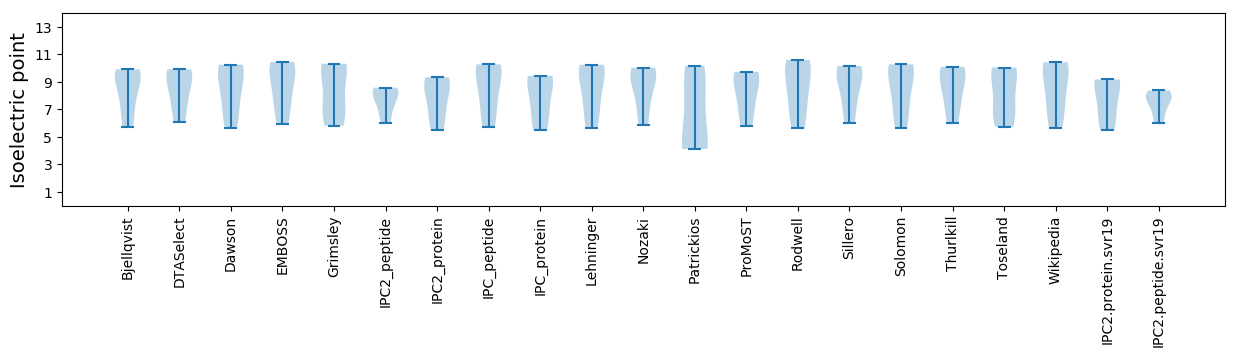
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q66125|2B_CMVQ Suppressor of silencing 2b OS=Cucumber mosaic virus (strain Q) OX=12310 GN=ORF2b PE=1 SV=1
MM1 pKa = 7.72ISPPPTFSFANLLNGSYY18 pKa = 10.84GVDD21 pKa = 3.02TPEE24 pKa = 3.53EE25 pKa = 4.2VEE27 pKa = 4.06RR28 pKa = 11.84VRR30 pKa = 11.84RR31 pKa = 11.84EE32 pKa = 3.74QRR34 pKa = 11.84EE35 pKa = 4.0DD36 pKa = 3.18AEE38 pKa = 4.31AALRR42 pKa = 11.84NYY44 pKa = 10.07KK45 pKa = 9.64PLPAVDD51 pKa = 3.8VSEE54 pKa = 4.26SVPRR58 pKa = 11.84DD59 pKa = 3.49EE60 pKa = 5.85PIVSQTVTAAPVTSVDD76 pKa = 3.5DD77 pKa = 4.2AFVSFGAEE85 pKa = 4.44DD86 pKa = 4.17YY87 pKa = 11.74LEE89 pKa = 4.32MSPSEE94 pKa = 3.96LLSAFEE100 pKa = 5.25LMVKK104 pKa = 9.47PLRR107 pKa = 11.84VGEE110 pKa = 4.19VLCSSFDD117 pKa = 3.29RR118 pKa = 11.84SLFISSVAMARR129 pKa = 11.84TLLLAPLTSTRR140 pKa = 11.84TLKK143 pKa = 10.69RR144 pKa = 11.84FEE146 pKa = 4.9DD147 pKa = 4.1LVAAIYY153 pKa = 10.48LKK155 pKa = 10.06TDD157 pKa = 4.17FFLEE161 pKa = 4.39DD162 pKa = 5.21DD163 pKa = 4.56GPQTDD168 pKa = 3.82VSQSDD173 pKa = 3.83VPGYY177 pKa = 10.1IFEE180 pKa = 5.44PGQHH184 pKa = 6.05SSGFEE189 pKa = 3.84PPPICAKK196 pKa = 10.32CDD198 pKa = 4.26LILYY202 pKa = 7.3QCPCFDD208 pKa = 4.27FNALRR213 pKa = 11.84EE214 pKa = 4.32SCAEE218 pKa = 3.78KK219 pKa = 10.15TFSHH223 pKa = 7.35DD224 pKa = 3.74YY225 pKa = 10.68VIEE228 pKa = 4.31GLDD231 pKa = 3.65GVIDD235 pKa = 3.91NATLLSNLGPFLLPVHH251 pKa = 6.63CSYY254 pKa = 11.74SKK256 pKa = 9.93TEE258 pKa = 4.45DD259 pKa = 3.45PDD261 pKa = 4.25FVVDD265 pKa = 4.49PSLARR270 pKa = 11.84PTDD273 pKa = 3.41RR274 pKa = 11.84VDD276 pKa = 3.14VHH278 pKa = 6.64VVQAVCDD285 pKa = 3.95TTLPTHH291 pKa = 6.64GNYY294 pKa = 10.09DD295 pKa = 3.56DD296 pKa = 5.53SFHH299 pKa = 6.76QVFVDD304 pKa = 3.45SADD307 pKa = 3.39YY308 pKa = 9.48STDD311 pKa = 3.1MDD313 pKa = 3.96HH314 pKa = 7.27VRR316 pKa = 11.84LRR318 pKa = 11.84QSDD321 pKa = 4.44LVAKK325 pKa = 10.26IPDD328 pKa = 4.31GGHH331 pKa = 6.04MLPVLNTGSGHH342 pKa = 5.29QRR344 pKa = 11.84VGTTKK349 pKa = 10.48EE350 pKa = 3.66VLTAIKK356 pKa = 9.97KK357 pKa = 10.04RR358 pKa = 11.84NADD361 pKa = 3.49VPEE364 pKa = 5.43LGDD367 pKa = 3.75SVNLSRR373 pKa = 11.84LSKK376 pKa = 10.62AVAEE380 pKa = 4.41RR381 pKa = 11.84FRR383 pKa = 11.84LSYY386 pKa = 10.7MNVDD390 pKa = 4.39ALAKK394 pKa = 10.03SNFVNVVSNFHH405 pKa = 7.84AYY407 pKa = 6.3MQKK410 pKa = 9.45WPSSGLSYY418 pKa = 11.44DD419 pKa = 4.67DD420 pKa = 5.94LPDD423 pKa = 3.54LHH425 pKa = 8.26AEE427 pKa = 3.82NLQFYY432 pKa = 10.96DD433 pKa = 4.19HH434 pKa = 6.9MIKK437 pKa = 10.37SDD439 pKa = 3.7VKK441 pKa = 10.7PVVTDD446 pKa = 3.18TLNVDD451 pKa = 3.69RR452 pKa = 11.84PVPATITFHH461 pKa = 7.02KK462 pKa = 9.73KK463 pKa = 8.81TITSQFSPLFISLFEE478 pKa = 4.05RR479 pKa = 11.84FQRR482 pKa = 11.84CLRR485 pKa = 11.84EE486 pKa = 4.05RR487 pKa = 11.84VVLPVGKK494 pKa = 9.96ISSLEE499 pKa = 3.79MTGFSVLNKK508 pKa = 9.86HH509 pKa = 6.15CLEE512 pKa = 4.71IDD514 pKa = 3.33LSKK517 pKa = 10.66FDD519 pKa = 3.64KK520 pKa = 11.1SQGEE524 pKa = 4.13FHH526 pKa = 7.92LMIQEE531 pKa = 5.03HH532 pKa = 6.55ILNDD536 pKa = 4.6LGCPAPITKK545 pKa = 8.63WWCDD549 pKa = 3.85FHH551 pKa = 8.08RR552 pKa = 11.84FSYY555 pKa = 10.65IKK557 pKa = 10.24DD558 pKa = 3.05KK559 pKa = 10.9RR560 pKa = 11.84AGVGMPISFQRR571 pKa = 11.84RR572 pKa = 11.84TGDD575 pKa = 2.78AFTYY579 pKa = 10.13FGNTIVTMAEE589 pKa = 4.01FAWCYY594 pKa = 10.05DD595 pKa = 2.91TDD597 pKa = 3.68QFDD600 pKa = 3.97RR601 pKa = 11.84LLFSGDD607 pKa = 3.51DD608 pKa = 3.31SLAFSKK614 pKa = 10.59LPPVGDD620 pKa = 3.47PSKK623 pKa = 10.39FTTLFNMEE631 pKa = 4.46AKK633 pKa = 10.42VMEE636 pKa = 4.39PAVPYY641 pKa = 9.94ICSKK645 pKa = 10.49FYY647 pKa = 11.44SLMSLVTRR655 pKa = 11.84FQSPTIRR662 pKa = 11.84EE663 pKa = 4.02IQRR666 pKa = 11.84LGTKK670 pKa = 10.02KK671 pKa = 10.19IPYY674 pKa = 9.41SDD676 pKa = 4.15NNDD679 pKa = 2.96FLFAHH684 pKa = 6.57FMSFVDD690 pKa = 3.62RR691 pKa = 11.84LKK693 pKa = 11.3FMDD696 pKa = 5.36RR697 pKa = 11.84MSQSCIDD704 pKa = 3.38QLSIFFEE711 pKa = 4.35LKK713 pKa = 9.87YY714 pKa = 10.59KK715 pKa = 10.81KK716 pKa = 10.27SGNEE720 pKa = 3.43AALVLGAFKK729 pKa = 10.8KK730 pKa = 9.51YY731 pKa = 7.93TANFNAYY738 pKa = 9.36KK739 pKa = 10.08EE740 pKa = 4.67LYY742 pKa = 10.2YY743 pKa = 10.53SDD745 pKa = 5.12RR746 pKa = 11.84QQCDD750 pKa = 3.15LVNTFCISEE759 pKa = 3.88FRR761 pKa = 11.84VIRR764 pKa = 11.84RR765 pKa = 11.84TTVKK769 pKa = 10.08KK770 pKa = 10.53KK771 pKa = 10.91KK772 pKa = 9.79NGCVDD777 pKa = 2.87SSGVDD782 pKa = 3.17RR783 pKa = 11.84RR784 pKa = 11.84PPLSQFAGGEE794 pKa = 4.1TSKK797 pKa = 10.06TKK799 pKa = 10.49VSRR802 pKa = 11.84QKK804 pKa = 10.15PASEE808 pKa = 4.46GLQKK812 pKa = 10.23SQRR815 pKa = 11.84EE816 pKa = 3.99SAIYY820 pKa = 10.56SEE822 pKa = 4.64TFPDD826 pKa = 3.42VTIPRR831 pKa = 11.84SRR833 pKa = 11.84SRR835 pKa = 11.84GLVSS839 pKa = 3.41
MM1 pKa = 7.72ISPPPTFSFANLLNGSYY18 pKa = 10.84GVDD21 pKa = 3.02TPEE24 pKa = 3.53EE25 pKa = 4.2VEE27 pKa = 4.06RR28 pKa = 11.84VRR30 pKa = 11.84RR31 pKa = 11.84EE32 pKa = 3.74QRR34 pKa = 11.84EE35 pKa = 4.0DD36 pKa = 3.18AEE38 pKa = 4.31AALRR42 pKa = 11.84NYY44 pKa = 10.07KK45 pKa = 9.64PLPAVDD51 pKa = 3.8VSEE54 pKa = 4.26SVPRR58 pKa = 11.84DD59 pKa = 3.49EE60 pKa = 5.85PIVSQTVTAAPVTSVDD76 pKa = 3.5DD77 pKa = 4.2AFVSFGAEE85 pKa = 4.44DD86 pKa = 4.17YY87 pKa = 11.74LEE89 pKa = 4.32MSPSEE94 pKa = 3.96LLSAFEE100 pKa = 5.25LMVKK104 pKa = 9.47PLRR107 pKa = 11.84VGEE110 pKa = 4.19VLCSSFDD117 pKa = 3.29RR118 pKa = 11.84SLFISSVAMARR129 pKa = 11.84TLLLAPLTSTRR140 pKa = 11.84TLKK143 pKa = 10.69RR144 pKa = 11.84FEE146 pKa = 4.9DD147 pKa = 4.1LVAAIYY153 pKa = 10.48LKK155 pKa = 10.06TDD157 pKa = 4.17FFLEE161 pKa = 4.39DD162 pKa = 5.21DD163 pKa = 4.56GPQTDD168 pKa = 3.82VSQSDD173 pKa = 3.83VPGYY177 pKa = 10.1IFEE180 pKa = 5.44PGQHH184 pKa = 6.05SSGFEE189 pKa = 3.84PPPICAKK196 pKa = 10.32CDD198 pKa = 4.26LILYY202 pKa = 7.3QCPCFDD208 pKa = 4.27FNALRR213 pKa = 11.84EE214 pKa = 4.32SCAEE218 pKa = 3.78KK219 pKa = 10.15TFSHH223 pKa = 7.35DD224 pKa = 3.74YY225 pKa = 10.68VIEE228 pKa = 4.31GLDD231 pKa = 3.65GVIDD235 pKa = 3.91NATLLSNLGPFLLPVHH251 pKa = 6.63CSYY254 pKa = 11.74SKK256 pKa = 9.93TEE258 pKa = 4.45DD259 pKa = 3.45PDD261 pKa = 4.25FVVDD265 pKa = 4.49PSLARR270 pKa = 11.84PTDD273 pKa = 3.41RR274 pKa = 11.84VDD276 pKa = 3.14VHH278 pKa = 6.64VVQAVCDD285 pKa = 3.95TTLPTHH291 pKa = 6.64GNYY294 pKa = 10.09DD295 pKa = 3.56DD296 pKa = 5.53SFHH299 pKa = 6.76QVFVDD304 pKa = 3.45SADD307 pKa = 3.39YY308 pKa = 9.48STDD311 pKa = 3.1MDD313 pKa = 3.96HH314 pKa = 7.27VRR316 pKa = 11.84LRR318 pKa = 11.84QSDD321 pKa = 4.44LVAKK325 pKa = 10.26IPDD328 pKa = 4.31GGHH331 pKa = 6.04MLPVLNTGSGHH342 pKa = 5.29QRR344 pKa = 11.84VGTTKK349 pKa = 10.48EE350 pKa = 3.66VLTAIKK356 pKa = 9.97KK357 pKa = 10.04RR358 pKa = 11.84NADD361 pKa = 3.49VPEE364 pKa = 5.43LGDD367 pKa = 3.75SVNLSRR373 pKa = 11.84LSKK376 pKa = 10.62AVAEE380 pKa = 4.41RR381 pKa = 11.84FRR383 pKa = 11.84LSYY386 pKa = 10.7MNVDD390 pKa = 4.39ALAKK394 pKa = 10.03SNFVNVVSNFHH405 pKa = 7.84AYY407 pKa = 6.3MQKK410 pKa = 9.45WPSSGLSYY418 pKa = 11.44DD419 pKa = 4.67DD420 pKa = 5.94LPDD423 pKa = 3.54LHH425 pKa = 8.26AEE427 pKa = 3.82NLQFYY432 pKa = 10.96DD433 pKa = 4.19HH434 pKa = 6.9MIKK437 pKa = 10.37SDD439 pKa = 3.7VKK441 pKa = 10.7PVVTDD446 pKa = 3.18TLNVDD451 pKa = 3.69RR452 pKa = 11.84PVPATITFHH461 pKa = 7.02KK462 pKa = 9.73KK463 pKa = 8.81TITSQFSPLFISLFEE478 pKa = 4.05RR479 pKa = 11.84FQRR482 pKa = 11.84CLRR485 pKa = 11.84EE486 pKa = 4.05RR487 pKa = 11.84VVLPVGKK494 pKa = 9.96ISSLEE499 pKa = 3.79MTGFSVLNKK508 pKa = 9.86HH509 pKa = 6.15CLEE512 pKa = 4.71IDD514 pKa = 3.33LSKK517 pKa = 10.66FDD519 pKa = 3.64KK520 pKa = 11.1SQGEE524 pKa = 4.13FHH526 pKa = 7.92LMIQEE531 pKa = 5.03HH532 pKa = 6.55ILNDD536 pKa = 4.6LGCPAPITKK545 pKa = 8.63WWCDD549 pKa = 3.85FHH551 pKa = 8.08RR552 pKa = 11.84FSYY555 pKa = 10.65IKK557 pKa = 10.24DD558 pKa = 3.05KK559 pKa = 10.9RR560 pKa = 11.84AGVGMPISFQRR571 pKa = 11.84RR572 pKa = 11.84TGDD575 pKa = 2.78AFTYY579 pKa = 10.13FGNTIVTMAEE589 pKa = 4.01FAWCYY594 pKa = 10.05DD595 pKa = 2.91TDD597 pKa = 3.68QFDD600 pKa = 3.97RR601 pKa = 11.84LLFSGDD607 pKa = 3.51DD608 pKa = 3.31SLAFSKK614 pKa = 10.59LPPVGDD620 pKa = 3.47PSKK623 pKa = 10.39FTTLFNMEE631 pKa = 4.46AKK633 pKa = 10.42VMEE636 pKa = 4.39PAVPYY641 pKa = 9.94ICSKK645 pKa = 10.49FYY647 pKa = 11.44SLMSLVTRR655 pKa = 11.84FQSPTIRR662 pKa = 11.84EE663 pKa = 4.02IQRR666 pKa = 11.84LGTKK670 pKa = 10.02KK671 pKa = 10.19IPYY674 pKa = 9.41SDD676 pKa = 4.15NNDD679 pKa = 2.96FLFAHH684 pKa = 6.57FMSFVDD690 pKa = 3.62RR691 pKa = 11.84LKK693 pKa = 11.3FMDD696 pKa = 5.36RR697 pKa = 11.84MSQSCIDD704 pKa = 3.38QLSIFFEE711 pKa = 4.35LKK713 pKa = 9.87YY714 pKa = 10.59KK715 pKa = 10.81KK716 pKa = 10.27SGNEE720 pKa = 3.43AALVLGAFKK729 pKa = 10.8KK730 pKa = 9.51YY731 pKa = 7.93TANFNAYY738 pKa = 9.36KK739 pKa = 10.08EE740 pKa = 4.67LYY742 pKa = 10.2YY743 pKa = 10.53SDD745 pKa = 5.12RR746 pKa = 11.84QQCDD750 pKa = 3.15LVNTFCISEE759 pKa = 3.88FRR761 pKa = 11.84VIRR764 pKa = 11.84RR765 pKa = 11.84TTVKK769 pKa = 10.08KK770 pKa = 10.53KK771 pKa = 10.91KK772 pKa = 9.79NGCVDD777 pKa = 2.87SSGVDD782 pKa = 3.17RR783 pKa = 11.84RR784 pKa = 11.84PPLSQFAGGEE794 pKa = 4.1TSKK797 pKa = 10.06TKK799 pKa = 10.49VSRR802 pKa = 11.84QKK804 pKa = 10.15PASEE808 pKa = 4.46GLQKK812 pKa = 10.23SQRR815 pKa = 11.84EE816 pKa = 3.99SAIYY820 pKa = 10.56SEE822 pKa = 4.64TFPDD826 pKa = 3.42VTIPRR831 pKa = 11.84SRR833 pKa = 11.84SRR835 pKa = 11.84GLVSS839 pKa = 3.41
Molecular weight: 94.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q83267|Q83267_CMVQ Capsid protein OS=Cucumber mosaic virus (strain Q) OX=12310 PE=3 SV=1
MM1 pKa = 7.95DD2 pKa = 5.07KK3 pKa = 10.83SGSPNASRR11 pKa = 11.84TSGRR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84RR19 pKa = 11.84GSRR22 pKa = 11.84SASGADD28 pKa = 2.93AGLRR32 pKa = 11.84ALTQQMLRR40 pKa = 11.84LNKK43 pKa = 8.8TLAIGRR49 pKa = 11.84PTLNHH54 pKa = 5.13QPSWVVKK61 pKa = 9.82AVNPGYY67 pKa = 8.78TFTSITLKK75 pKa = 10.54PPEE78 pKa = 4.38IEE80 pKa = 4.22KK81 pKa = 10.65GSCFGRR87 pKa = 11.84RR88 pKa = 11.84LSLPDD93 pKa = 3.75SVTDD97 pKa = 3.63YY98 pKa = 11.42DD99 pKa = 3.77KK100 pKa = 11.56KK101 pKa = 10.74LVSRR105 pKa = 11.84IQIRR109 pKa = 11.84INPLPKK115 pKa = 9.55FDD117 pKa = 3.58STVWVTVRR125 pKa = 11.84KK126 pKa = 9.96VPSSSDD132 pKa = 3.29LSVAAISAMFGDD144 pKa = 4.59GNSPVLVYY152 pKa = 9.68QYY154 pKa = 11.09AASGVQANNKK164 pKa = 9.16LLYY167 pKa = 10.29DD168 pKa = 4.29LSEE171 pKa = 3.92MRR173 pKa = 11.84ADD175 pKa = 3.14IGDD178 pKa = 3.55MRR180 pKa = 11.84KK181 pKa = 9.47YY182 pKa = 10.88AVLVYY187 pKa = 10.91SKK189 pKa = 10.91DD190 pKa = 3.64DD191 pKa = 3.61KK192 pKa = 11.49LEE194 pKa = 3.97KK195 pKa = 10.77DD196 pKa = 4.07EE197 pKa = 4.32IVFMSTSSINEE208 pKa = 4.08FLSHH212 pKa = 6.06GCSRR216 pKa = 11.84LSPCVYY222 pKa = 10.17RR223 pKa = 11.84RR224 pKa = 11.84PKK226 pKa = 8.57TLNYY230 pKa = 8.95TLNRR234 pKa = 11.84EE235 pKa = 4.37CC236 pKa = 5.61
MM1 pKa = 7.95DD2 pKa = 5.07KK3 pKa = 10.83SGSPNASRR11 pKa = 11.84TSGRR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84RR19 pKa = 11.84GSRR22 pKa = 11.84SASGADD28 pKa = 2.93AGLRR32 pKa = 11.84ALTQQMLRR40 pKa = 11.84LNKK43 pKa = 8.8TLAIGRR49 pKa = 11.84PTLNHH54 pKa = 5.13QPSWVVKK61 pKa = 9.82AVNPGYY67 pKa = 8.78TFTSITLKK75 pKa = 10.54PPEE78 pKa = 4.38IEE80 pKa = 4.22KK81 pKa = 10.65GSCFGRR87 pKa = 11.84RR88 pKa = 11.84LSLPDD93 pKa = 3.75SVTDD97 pKa = 3.63YY98 pKa = 11.42DD99 pKa = 3.77KK100 pKa = 11.56KK101 pKa = 10.74LVSRR105 pKa = 11.84IQIRR109 pKa = 11.84INPLPKK115 pKa = 9.55FDD117 pKa = 3.58STVWVTVRR125 pKa = 11.84KK126 pKa = 9.96VPSSSDD132 pKa = 3.29LSVAAISAMFGDD144 pKa = 4.59GNSPVLVYY152 pKa = 9.68QYY154 pKa = 11.09AASGVQANNKK164 pKa = 9.16LLYY167 pKa = 10.29DD168 pKa = 4.29LSEE171 pKa = 3.92MRR173 pKa = 11.84ADD175 pKa = 3.14IGDD178 pKa = 3.55MRR180 pKa = 11.84KK181 pKa = 9.47YY182 pKa = 10.88AVLVYY187 pKa = 10.91SKK189 pKa = 10.91DD190 pKa = 3.64DD191 pKa = 3.61KK192 pKa = 11.49LEE194 pKa = 3.97KK195 pKa = 10.77DD196 pKa = 4.07EE197 pKa = 4.32IVFMSTSSINEE208 pKa = 4.08FLSHH212 pKa = 6.06GCSRR216 pKa = 11.84LSPCVYY222 pKa = 10.17RR223 pKa = 11.84RR224 pKa = 11.84PKK226 pKa = 8.57TLNYY230 pKa = 8.95TLNRR234 pKa = 11.84EE235 pKa = 4.37CC236 pKa = 5.61
Molecular weight: 26.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2663 |
100 |
991 |
443.8 |
49.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.834 ± 0.45 | 2.103 ± 0.355 |
6.647 ± 0.504 | 4.732 ± 0.463 |
4.957 ± 0.868 | 5.407 ± 0.367 |
2.253 ± 0.304 | 4.318 ± 0.349 |
5.745 ± 0.382 | 9.05 ± 0.167 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.591 ± 0.237 | 3.567 ± 0.448 |
4.957 ± 0.723 | 3.154 ± 0.236 |
6.234 ± 0.837 | 9.688 ± 0.825 |
5.971 ± 0.389 | 7.773 ± 0.276 |
0.901 ± 0.208 | 3.117 ± 0.162 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
