
Octopus bimaculoides (California two-spotted octopus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Mollusca; Cephalopoda; Coleoidea; Octopodiformes; Octopoda; Incirrata; Octopodidae; Octopus
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins
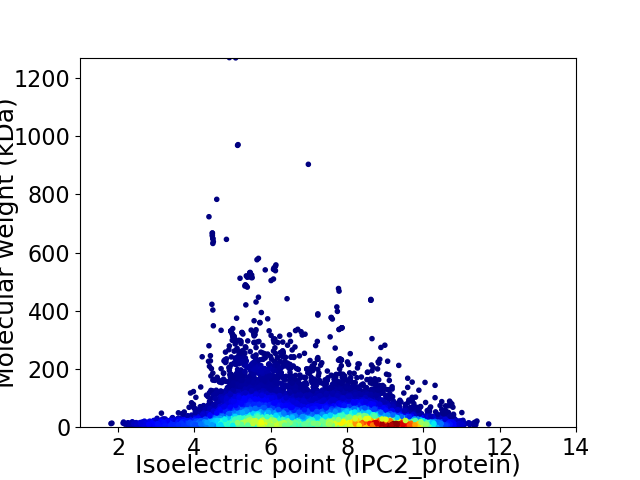
Virtual 2D-PAGE plot for 36239 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0L8G9Y1|A0A0L8G9Y1_OCTBM Uncharacterized protein OS=Octopus bimaculoides OX=37653 GN=OCBIM_22037958mg PE=4 SV=1
MM1 pKa = 7.73ADD3 pKa = 3.17EE4 pKa = 5.96GINNMTQPDD13 pKa = 4.13SVGNLLGDD21 pKa = 3.65SHH23 pKa = 8.04LGNLDD28 pKa = 3.72NYY30 pKa = 10.64SSSAFQSEE38 pKa = 4.32PLQPQPSAPHH48 pKa = 6.72QMMSSGSISSSNPLDD63 pKa = 4.12DD64 pKa = 5.91DD65 pKa = 4.58FGFDD69 pKa = 4.02SHH71 pKa = 6.28ITSSNTVNPSSNTTSSGSNNSAQSSSSYY99 pKa = 10.76TPGEE103 pKa = 4.5LISNTYY109 pKa = 10.64NPVDD113 pKa = 3.51HH114 pKa = 7.63DD115 pKa = 4.67SNYY118 pKa = 9.97NASRR122 pKa = 11.84VQIATHH128 pKa = 5.91DD129 pKa = 3.43TGEE132 pKa = 4.24NFNLDD137 pKa = 3.21NSNYY141 pKa = 9.91GMSDD145 pKa = 3.34SSPPPPLSQYY155 pKa = 7.42QQHH158 pKa = 6.61HH159 pKa = 6.54LNEE162 pKa = 4.54PSSKK166 pKa = 10.14VDD168 pKa = 3.49EE169 pKa = 4.49VDD171 pKa = 4.82SGDD174 pKa = 4.06SDD176 pKa = 4.07SDD178 pKa = 3.84DD179 pKa = 3.79YY180 pKa = 11.88EE181 pKa = 6.24KK182 pKa = 11.22YY183 pKa = 10.85DD184 pKa = 3.42HH185 pKa = 7.6DD186 pKa = 4.38SLVRR190 pKa = 11.84GQTVLTSKK198 pKa = 10.56HH199 pKa = 6.8SEE201 pKa = 4.2DD202 pKa = 3.64QQHH205 pKa = 6.06SQDD208 pKa = 4.22EE209 pKa = 4.43YY210 pKa = 11.66DD211 pKa = 5.67DD212 pKa = 4.84EE213 pKa = 7.61DD214 pKa = 6.16DD215 pKa = 5.24DD216 pKa = 5.65GEE218 pKa = 4.48SSLIEE223 pKa = 4.06YY224 pKa = 10.81NDD226 pKa = 3.7IEE228 pKa = 4.91DD229 pKa = 5.1DD230 pKa = 3.82EE231 pKa = 5.3VDD233 pKa = 3.79VVKK236 pKa = 11.18DD237 pKa = 3.34NDD239 pKa = 3.72FVYY242 pKa = 9.71STSAVADD249 pKa = 3.54SANAVDD255 pKa = 4.86KK256 pKa = 11.36FPDD259 pKa = 3.58NVQAPQLSPLDD270 pKa = 4.03SSLSPQTSPTHH281 pKa = 4.76QAPFHH286 pKa = 6.58SEE288 pKa = 3.48IPSFGSSEE296 pKa = 4.27ANDD299 pKa = 3.36VATISQSDD307 pKa = 3.51EE308 pKa = 4.15GKK310 pKa = 10.65VVDD313 pKa = 5.04SPSFMPSDD321 pKa = 3.61HH322 pKa = 7.15FITSTSDD329 pKa = 2.71AGTPSPSSVPVTSFGYY345 pKa = 10.42DD346 pKa = 3.32DD347 pKa = 4.36PLKK350 pKa = 10.76LSGVASSVEE359 pKa = 3.98TAAPSGQFMNDD370 pKa = 3.36LAPEE374 pKa = 4.31VIGGNDD380 pKa = 3.5GVEE383 pKa = 4.3VAGSEE388 pKa = 4.24NDD390 pKa = 3.96DD391 pKa = 4.02ADD393 pKa = 3.76SSDD396 pKa = 3.6EE397 pKa = 3.96QRR399 pKa = 11.84CEE401 pKa = 3.95IPAHH405 pKa = 6.48SSLSMSTPVVASANPSFSQPKK426 pKa = 8.28STSPPISDD434 pKa = 3.72PQLSVLNSDD443 pKa = 4.93PYY445 pKa = 10.49CTTIKK450 pKa = 10.08HH451 pKa = 4.98QQPSTFYY458 pKa = 8.3NTQSINEE465 pKa = 4.04NRR467 pKa = 11.84FSSEE471 pKa = 3.95PDD473 pKa = 3.11SSSEE477 pKa = 3.9PTVQTGPNFTEE488 pKa = 4.66PKK490 pKa = 10.16
MM1 pKa = 7.73ADD3 pKa = 3.17EE4 pKa = 5.96GINNMTQPDD13 pKa = 4.13SVGNLLGDD21 pKa = 3.65SHH23 pKa = 8.04LGNLDD28 pKa = 3.72NYY30 pKa = 10.64SSSAFQSEE38 pKa = 4.32PLQPQPSAPHH48 pKa = 6.72QMMSSGSISSSNPLDD63 pKa = 4.12DD64 pKa = 5.91DD65 pKa = 4.58FGFDD69 pKa = 4.02SHH71 pKa = 6.28ITSSNTVNPSSNTTSSGSNNSAQSSSSYY99 pKa = 10.76TPGEE103 pKa = 4.5LISNTYY109 pKa = 10.64NPVDD113 pKa = 3.51HH114 pKa = 7.63DD115 pKa = 4.67SNYY118 pKa = 9.97NASRR122 pKa = 11.84VQIATHH128 pKa = 5.91DD129 pKa = 3.43TGEE132 pKa = 4.24NFNLDD137 pKa = 3.21NSNYY141 pKa = 9.91GMSDD145 pKa = 3.34SSPPPPLSQYY155 pKa = 7.42QQHH158 pKa = 6.61HH159 pKa = 6.54LNEE162 pKa = 4.54PSSKK166 pKa = 10.14VDD168 pKa = 3.49EE169 pKa = 4.49VDD171 pKa = 4.82SGDD174 pKa = 4.06SDD176 pKa = 4.07SDD178 pKa = 3.84DD179 pKa = 3.79YY180 pKa = 11.88EE181 pKa = 6.24KK182 pKa = 11.22YY183 pKa = 10.85DD184 pKa = 3.42HH185 pKa = 7.6DD186 pKa = 4.38SLVRR190 pKa = 11.84GQTVLTSKK198 pKa = 10.56HH199 pKa = 6.8SEE201 pKa = 4.2DD202 pKa = 3.64QQHH205 pKa = 6.06SQDD208 pKa = 4.22EE209 pKa = 4.43YY210 pKa = 11.66DD211 pKa = 5.67DD212 pKa = 4.84EE213 pKa = 7.61DD214 pKa = 6.16DD215 pKa = 5.24DD216 pKa = 5.65GEE218 pKa = 4.48SSLIEE223 pKa = 4.06YY224 pKa = 10.81NDD226 pKa = 3.7IEE228 pKa = 4.91DD229 pKa = 5.1DD230 pKa = 3.82EE231 pKa = 5.3VDD233 pKa = 3.79VVKK236 pKa = 11.18DD237 pKa = 3.34NDD239 pKa = 3.72FVYY242 pKa = 9.71STSAVADD249 pKa = 3.54SANAVDD255 pKa = 4.86KK256 pKa = 11.36FPDD259 pKa = 3.58NVQAPQLSPLDD270 pKa = 4.03SSLSPQTSPTHH281 pKa = 4.76QAPFHH286 pKa = 6.58SEE288 pKa = 3.48IPSFGSSEE296 pKa = 4.27ANDD299 pKa = 3.36VATISQSDD307 pKa = 3.51EE308 pKa = 4.15GKK310 pKa = 10.65VVDD313 pKa = 5.04SPSFMPSDD321 pKa = 3.61HH322 pKa = 7.15FITSTSDD329 pKa = 2.71AGTPSPSSVPVTSFGYY345 pKa = 10.42DD346 pKa = 3.32DD347 pKa = 4.36PLKK350 pKa = 10.76LSGVASSVEE359 pKa = 3.98TAAPSGQFMNDD370 pKa = 3.36LAPEE374 pKa = 4.31VIGGNDD380 pKa = 3.5GVEE383 pKa = 4.3VAGSEE388 pKa = 4.24NDD390 pKa = 3.96DD391 pKa = 4.02ADD393 pKa = 3.76SSDD396 pKa = 3.6EE397 pKa = 3.96QRR399 pKa = 11.84CEE401 pKa = 3.95IPAHH405 pKa = 6.48SSLSMSTPVVASANPSFSQPKK426 pKa = 8.28STSPPISDD434 pKa = 3.72PQLSVLNSDD443 pKa = 4.93PYY445 pKa = 10.49CTTIKK450 pKa = 10.08HH451 pKa = 4.98QQPSTFYY458 pKa = 8.3NTQSINEE465 pKa = 4.04NRR467 pKa = 11.84FSSEE471 pKa = 3.95PDD473 pKa = 3.11SSSEE477 pKa = 3.9PTVQTGPNFTEE488 pKa = 4.66PKK490 pKa = 10.16
Molecular weight: 52.25 kDa
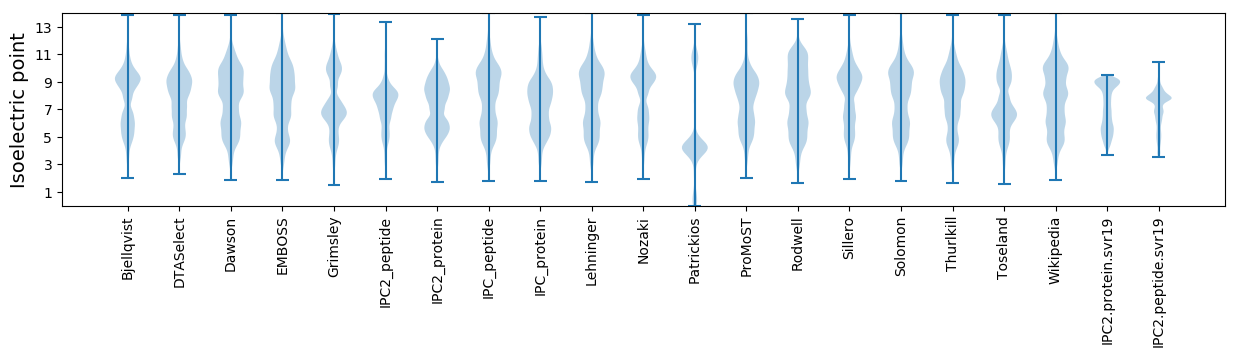
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L8GMG5|A0A0L8GMG5_OCTBM Chitin-binding type-4 domain-containing protein (Fragment) OS=Octopus bimaculoides OX=37653 GN=OCBIM_22031095mg PE=4 SV=1
EEE2 pKa = 4.34EE3 pKa = 3.99KK4 pKa = 9.69AKKK7 pKa = 8.33RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84WRR27 pKa = 11.84RR28 pKa = 11.84WRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KKK38 pKa = 7.93RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84WRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84KKK79 pKa = 9.13EE80 pKa = 3.49SMKKK84 pKa = 10.95NDDD87 pKa = 2.92VAISKKK93 pKa = 8.68K
EEE2 pKa = 4.34EE3 pKa = 3.99KK4 pKa = 9.69AKKK7 pKa = 8.33RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84WRR27 pKa = 11.84RR28 pKa = 11.84WRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KKK38 pKa = 7.93RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84WRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84KKK79 pKa = 9.13EE80 pKa = 3.49SMKKK84 pKa = 10.95NDDD87 pKa = 2.92VAISKKK93 pKa = 8.68K
Molecular weight: 13.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
12291891 |
8 |
11437 |
339.2 |
38.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.069 ± 0.014 | 2.44 ± 0.016 |
5.42 ± 0.018 | 6.562 ± 0.027 |
3.911 ± 0.014 | 4.945 ± 0.017 |
3.066 ± 0.015 | 5.969 ± 0.018 |
7.152 ± 0.021 | 8.581 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.274 ± 0.007 | 5.44 ± 0.014 |
4.568 ± 0.016 | 4.309 ± 0.019 |
4.905 ± 0.016 | 9.067 ± 0.029 |
6.421 ± 0.026 | 5.803 ± 0.013 |
0.934 ± 0.005 | 3.154 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
