
Fly associated circular virus 6
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.38
Get precalculated fractions of proteins
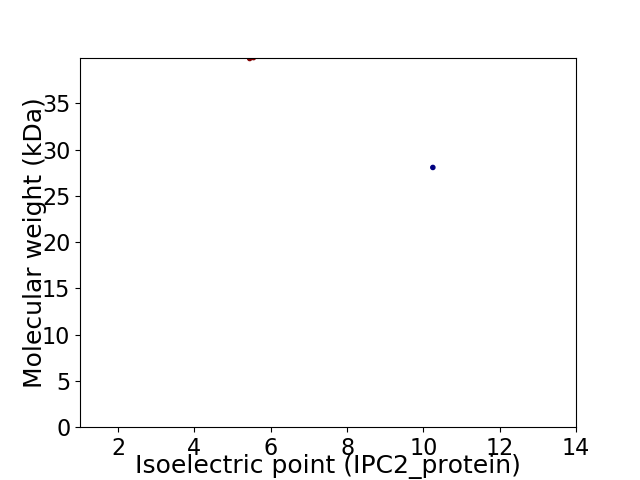
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPC5|A0A346BPC5_9VIRU Putative capsid protein OS=Fly associated circular virus 6 OX=2293286 PE=4 SV=1
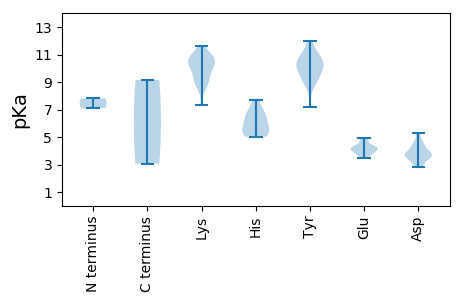
MM1 pKa = 7.82AGGARR6 pKa = 11.84NWCGTLQLEE15 pKa = 4.92DD16 pKa = 5.18DD17 pKa = 4.49DD18 pKa = 4.64WDD20 pKa = 4.52AEE22 pKa = 4.34TWLEE26 pKa = 4.24SLVRR30 pKa = 11.84DD31 pKa = 4.13EE32 pKa = 3.98KK33 pKa = 11.21AKK35 pKa = 10.74YY36 pKa = 9.99VVGQIEE42 pKa = 4.26RR43 pKa = 11.84GHH45 pKa = 5.12EE46 pKa = 3.97TEE48 pKa = 4.29HH49 pKa = 6.64AHH51 pKa = 5.82LQFYY55 pKa = 7.16VQCLSQKK62 pKa = 10.26NLVWMKK68 pKa = 11.01RR69 pKa = 11.84NINDD73 pKa = 3.47KK74 pKa = 10.9AHH76 pKa = 6.63WEE78 pKa = 4.22TAKK81 pKa = 9.75GTAKK85 pKa = 10.44HH86 pKa = 5.41NRR88 pKa = 11.84AYY90 pKa = 8.9CTKK93 pKa = 9.0EE94 pKa = 3.74DD95 pKa = 3.63SRR97 pKa = 11.84VRR99 pKa = 11.84GPWEE103 pKa = 3.8FGEE106 pKa = 4.72IVQQGQRR113 pKa = 11.84EE114 pKa = 4.24GLEE117 pKa = 4.16SAIKK121 pKa = 9.09MVKK124 pKa = 9.96EE125 pKa = 3.73GAPLKK130 pKa = 10.73DD131 pKa = 2.96IALEE135 pKa = 3.92QSVVWVHH142 pKa = 5.28HH143 pKa = 6.15HH144 pKa = 6.79RR145 pKa = 11.84GLMDD149 pKa = 3.74LRR151 pKa = 11.84KK152 pKa = 9.54QLGLEE157 pKa = 3.53ADD159 pKa = 3.36RR160 pKa = 11.84RR161 pKa = 11.84YY162 pKa = 10.55FGVDD166 pKa = 3.76GPEE169 pKa = 4.32VWCLWGPSGTGKK181 pKa = 10.36SRR183 pKa = 11.84WVNAKK188 pKa = 9.52WPDD191 pKa = 3.85AFWKK195 pKa = 10.38IPNEE199 pKa = 4.1KK200 pKa = 8.78WWDD203 pKa = 3.68GYY205 pKa = 9.32TGQEE209 pKa = 4.07TVILDD214 pKa = 3.71DD215 pKa = 5.26FKK217 pKa = 11.64DD218 pKa = 3.41GDD220 pKa = 3.81MRR222 pKa = 11.84LGDD225 pKa = 4.32LQRR228 pKa = 11.84LLDD231 pKa = 4.47WYY233 pKa = 9.17PLWVEE238 pKa = 4.35VKK240 pKa = 10.63GGTIPMLAKK249 pKa = 10.18RR250 pKa = 11.84YY251 pKa = 9.76VFTCNTHH258 pKa = 5.62PSSWYY263 pKa = 10.04RR264 pKa = 11.84KK265 pKa = 8.75ADD267 pKa = 3.33PHH269 pKa = 5.02RR270 pKa = 11.84TIMRR274 pKa = 11.84RR275 pKa = 11.84FDD277 pKa = 3.06EE278 pKa = 4.67FAAQKK283 pKa = 10.58GRR285 pKa = 11.84LIMCEE290 pKa = 3.74TDD292 pKa = 2.79DD293 pKa = 4.3WEE295 pKa = 4.88INLEE299 pKa = 4.13TRR301 pKa = 11.84LGDD304 pKa = 3.76LGNPQTPEE312 pKa = 3.68VGGNTSTPTSGAPLRR327 pKa = 11.84VPEE330 pKa = 4.11NSEE333 pKa = 4.95DD334 pKa = 3.51IMIPSPMDD342 pKa = 3.71FYY344 pKa = 12.01VV345 pKa = 3.06
MM1 pKa = 7.82AGGARR6 pKa = 11.84NWCGTLQLEE15 pKa = 4.92DD16 pKa = 5.18DD17 pKa = 4.49DD18 pKa = 4.64WDD20 pKa = 4.52AEE22 pKa = 4.34TWLEE26 pKa = 4.24SLVRR30 pKa = 11.84DD31 pKa = 4.13EE32 pKa = 3.98KK33 pKa = 11.21AKK35 pKa = 10.74YY36 pKa = 9.99VVGQIEE42 pKa = 4.26RR43 pKa = 11.84GHH45 pKa = 5.12EE46 pKa = 3.97TEE48 pKa = 4.29HH49 pKa = 6.64AHH51 pKa = 5.82LQFYY55 pKa = 7.16VQCLSQKK62 pKa = 10.26NLVWMKK68 pKa = 11.01RR69 pKa = 11.84NINDD73 pKa = 3.47KK74 pKa = 10.9AHH76 pKa = 6.63WEE78 pKa = 4.22TAKK81 pKa = 9.75GTAKK85 pKa = 10.44HH86 pKa = 5.41NRR88 pKa = 11.84AYY90 pKa = 8.9CTKK93 pKa = 9.0EE94 pKa = 3.74DD95 pKa = 3.63SRR97 pKa = 11.84VRR99 pKa = 11.84GPWEE103 pKa = 3.8FGEE106 pKa = 4.72IVQQGQRR113 pKa = 11.84EE114 pKa = 4.24GLEE117 pKa = 4.16SAIKK121 pKa = 9.09MVKK124 pKa = 9.96EE125 pKa = 3.73GAPLKK130 pKa = 10.73DD131 pKa = 2.96IALEE135 pKa = 3.92QSVVWVHH142 pKa = 5.28HH143 pKa = 6.15HH144 pKa = 6.79RR145 pKa = 11.84GLMDD149 pKa = 3.74LRR151 pKa = 11.84KK152 pKa = 9.54QLGLEE157 pKa = 3.53ADD159 pKa = 3.36RR160 pKa = 11.84RR161 pKa = 11.84YY162 pKa = 10.55FGVDD166 pKa = 3.76GPEE169 pKa = 4.32VWCLWGPSGTGKK181 pKa = 10.36SRR183 pKa = 11.84WVNAKK188 pKa = 9.52WPDD191 pKa = 3.85AFWKK195 pKa = 10.38IPNEE199 pKa = 4.1KK200 pKa = 8.78WWDD203 pKa = 3.68GYY205 pKa = 9.32TGQEE209 pKa = 4.07TVILDD214 pKa = 3.71DD215 pKa = 5.26FKK217 pKa = 11.64DD218 pKa = 3.41GDD220 pKa = 3.81MRR222 pKa = 11.84LGDD225 pKa = 4.32LQRR228 pKa = 11.84LLDD231 pKa = 4.47WYY233 pKa = 9.17PLWVEE238 pKa = 4.35VKK240 pKa = 10.63GGTIPMLAKK249 pKa = 10.18RR250 pKa = 11.84YY251 pKa = 9.76VFTCNTHH258 pKa = 5.62PSSWYY263 pKa = 10.04RR264 pKa = 11.84KK265 pKa = 8.75ADD267 pKa = 3.33PHH269 pKa = 5.02RR270 pKa = 11.84TIMRR274 pKa = 11.84RR275 pKa = 11.84FDD277 pKa = 3.06EE278 pKa = 4.67FAAQKK283 pKa = 10.58GRR285 pKa = 11.84LIMCEE290 pKa = 3.74TDD292 pKa = 2.79DD293 pKa = 4.3WEE295 pKa = 4.88INLEE299 pKa = 4.13TRR301 pKa = 11.84LGDD304 pKa = 3.76LGNPQTPEE312 pKa = 3.68VGGNTSTPTSGAPLRR327 pKa = 11.84VPEE330 pKa = 4.11NSEE333 pKa = 4.95DD334 pKa = 3.51IMIPSPMDD342 pKa = 3.71FYY344 pKa = 12.01VV345 pKa = 3.06
Molecular weight: 39.84 kDa
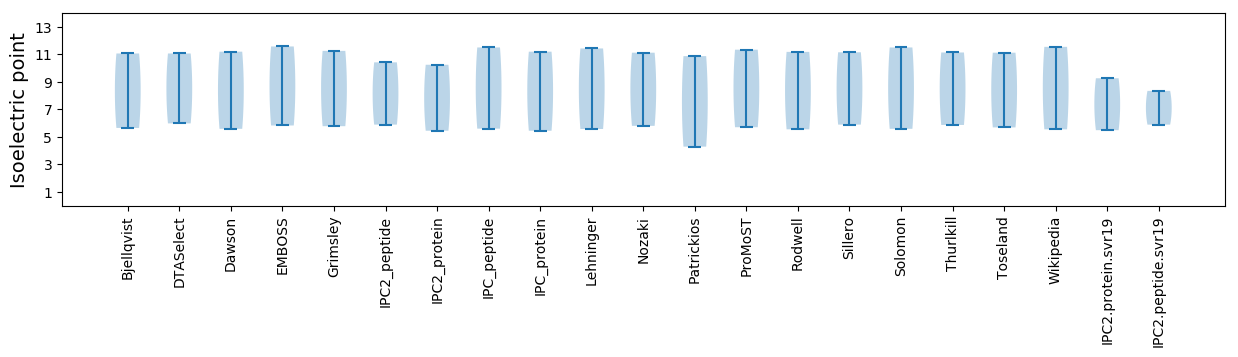
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPC5|A0A346BPC5_9VIRU Putative capsid protein OS=Fly associated circular virus 6 OX=2293286 PE=4 SV=1
MM1 pKa = 7.09AWSRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84TRR9 pKa = 11.84FQRR12 pKa = 11.84QRR14 pKa = 11.84PRR16 pKa = 11.84RR17 pKa = 11.84LRR19 pKa = 11.84RR20 pKa = 11.84ARR22 pKa = 11.84RR23 pKa = 11.84LTRR26 pKa = 11.84TRR28 pKa = 11.84FRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 9.02RR33 pKa = 11.84PRR35 pKa = 11.84GGGTRR40 pKa = 11.84SATVKK45 pKa = 9.47LTAEE49 pKa = 3.99TTYY52 pKa = 11.35APWQGPLTNVQAGWKK67 pKa = 8.59PVAFTPQNLQGFNDD81 pKa = 3.83YY82 pKa = 11.07QSVYY86 pKa = 10.44SQFRR90 pKa = 11.84VKK92 pKa = 10.59KK93 pKa = 9.84CVVKK97 pKa = 10.68INRR100 pKa = 11.84GPDD103 pKa = 3.1TNLVYY108 pKa = 10.78LVVPSRR114 pKa = 11.84AFAQTAAPSSSIANSWLAYY133 pKa = 9.96VPPQTEE139 pKa = 3.47QALRR143 pKa = 11.84QTRR146 pKa = 11.84WQKK149 pKa = 11.06EE150 pKa = 4.06LMPSTTAGKK159 pKa = 10.23VRR161 pKa = 11.84FGFHH165 pKa = 7.7PYY167 pKa = 10.38TMITAMGPIAQGQSINFTRR186 pKa = 11.84VWNLNKK192 pKa = 7.35WTPMSWALAQYY203 pKa = 9.28PMYY206 pKa = 10.18MYY208 pKa = 10.93GPYY211 pKa = 10.51LCLNNALTNADD222 pKa = 3.92PDD224 pKa = 3.8LTTSVSCIIEE234 pKa = 4.47CYY236 pKa = 10.22FQFKK240 pKa = 9.02GQKK243 pKa = 9.15
MM1 pKa = 7.09AWSRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84TRR9 pKa = 11.84FQRR12 pKa = 11.84QRR14 pKa = 11.84PRR16 pKa = 11.84RR17 pKa = 11.84LRR19 pKa = 11.84RR20 pKa = 11.84ARR22 pKa = 11.84RR23 pKa = 11.84LTRR26 pKa = 11.84TRR28 pKa = 11.84FRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 9.02RR33 pKa = 11.84PRR35 pKa = 11.84GGGTRR40 pKa = 11.84SATVKK45 pKa = 9.47LTAEE49 pKa = 3.99TTYY52 pKa = 11.35APWQGPLTNVQAGWKK67 pKa = 8.59PVAFTPQNLQGFNDD81 pKa = 3.83YY82 pKa = 11.07QSVYY86 pKa = 10.44SQFRR90 pKa = 11.84VKK92 pKa = 10.59KK93 pKa = 9.84CVVKK97 pKa = 10.68INRR100 pKa = 11.84GPDD103 pKa = 3.1TNLVYY108 pKa = 10.78LVVPSRR114 pKa = 11.84AFAQTAAPSSSIANSWLAYY133 pKa = 9.96VPPQTEE139 pKa = 3.47QALRR143 pKa = 11.84QTRR146 pKa = 11.84WQKK149 pKa = 11.06EE150 pKa = 4.06LMPSTTAGKK159 pKa = 10.23VRR161 pKa = 11.84FGFHH165 pKa = 7.7PYY167 pKa = 10.38TMITAMGPIAQGQSINFTRR186 pKa = 11.84VWNLNKK192 pKa = 7.35WTPMSWALAQYY203 pKa = 9.28PMYY206 pKa = 10.18MYY208 pKa = 10.93GPYY211 pKa = 10.51LCLNNALTNADD222 pKa = 3.92PDD224 pKa = 3.8LTTSVSCIIEE234 pKa = 4.47CYY236 pKa = 10.22FQFKK240 pKa = 9.02GQKK243 pKa = 9.15
Molecular weight: 28.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
588 |
243 |
345 |
294.0 |
33.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.973 ± 1.104 | 1.701 ± 0.036 |
5.102 ± 2.285 | 5.272 ± 2.397 |
3.401 ± 0.744 | 7.313 ± 1.298 |
1.871 ± 0.965 | 3.401 ± 0.344 |
5.272 ± 0.765 | 7.313 ± 0.482 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.891 ± 0.007 | 4.082 ± 0.566 |
5.952 ± 0.962 | 5.442 ± 1.299 |
8.333 ± 1.564 | 4.592 ± 0.773 |
7.143 ± 1.535 | 5.952 ± 0.126 |
4.422 ± 0.747 | 3.571 ± 0.904 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
