
Beihai tombus-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
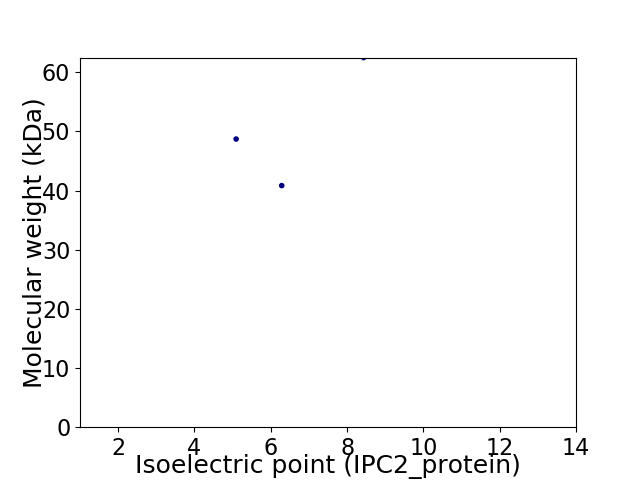
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGT8|A0A1L3KGT8_9VIRU RNA-directed RNA polymerase OS=Beihai tombus-like virus 4 OX=1922725 PE=4 SV=1
MM1 pKa = 7.54AVGLGWRR8 pKa = 11.84VIACNRR14 pKa = 11.84LRR16 pKa = 11.84YY17 pKa = 9.65LCLLNFCLTRR27 pKa = 11.84NTNNKK32 pKa = 9.61RR33 pKa = 11.84NTNRR37 pKa = 11.84SQRR40 pKa = 11.84VAAPVATGLRR50 pKa = 11.84TSGGRR55 pKa = 11.84PQTTPAGAGAIRR67 pKa = 11.84VRR69 pKa = 11.84HH70 pKa = 5.85RR71 pKa = 11.84EE72 pKa = 3.84FIKK75 pKa = 10.98NITSEE80 pKa = 4.14HH81 pKa = 5.28TFEE84 pKa = 5.38NGVLEE89 pKa = 4.29LGINAGDD96 pKa = 3.45TEE98 pKa = 4.34MFPWLSNIANGYY110 pKa = 8.18EE111 pKa = 4.15RR112 pKa = 11.84YY113 pKa = 9.4CVNSMSISYY122 pKa = 10.14EE123 pKa = 3.86PFVSTTEE130 pKa = 3.83SGAVIMQVDD139 pKa = 4.58YY140 pKa = 11.38DD141 pKa = 4.22PADD144 pKa = 3.8EE145 pKa = 4.55PPISKK150 pKa = 9.74SHH152 pKa = 5.77MLNSLGATRR161 pKa = 11.84SAVWMRR167 pKa = 11.84SAMALNRR174 pKa = 11.84KK175 pKa = 7.73EE176 pKa = 4.86LSYY179 pKa = 11.33DD180 pKa = 2.93DD181 pKa = 4.41HH182 pKa = 8.57LFVRR186 pKa = 11.84HH187 pKa = 5.7AVRR190 pKa = 11.84AGFSEE195 pKa = 3.79NLKK198 pKa = 10.56LYY200 pKa = 10.67DD201 pKa = 3.43VGTVFIALTDD211 pKa = 3.77VPAPTTLSYY220 pKa = 11.58GEE222 pKa = 4.06VWVTYY227 pKa = 10.08DD228 pKa = 2.94ITLMVPAFHH237 pKa = 6.62KK238 pKa = 10.59QEE240 pKa = 4.23PNAAEE245 pKa = 4.22SKK247 pKa = 10.45VNQSNYY253 pKa = 9.85FDD255 pKa = 3.49ILGTISSDD263 pKa = 3.51RR264 pKa = 11.84PNTMLQGSSINFTTDD279 pKa = 2.37SNNAGEE285 pKa = 4.27CAITFQEE292 pKa = 4.68PFTGLLYY299 pKa = 10.86LEE301 pKa = 4.14QSGYY305 pKa = 11.31AEE307 pKa = 4.82DD308 pKa = 5.04SITHH312 pKa = 6.75LEE314 pKa = 4.32LEE316 pKa = 4.62PTTTPADD323 pKa = 3.13NWITKK328 pKa = 9.03LARR331 pKa = 11.84LGGGVIDD338 pKa = 6.63YY339 pKa = 10.91ILLDD343 pKa = 4.05NKK345 pKa = 9.24WKK347 pKa = 10.59YY348 pKa = 10.56LIEE351 pKa = 4.22VVADD355 pKa = 3.7AGEE358 pKa = 4.56SIVFDD363 pKa = 4.19ALAVGAGDD371 pKa = 3.57LTTWIGEE378 pKa = 4.16TAMALSPYY386 pKa = 10.46AEE388 pKa = 4.22ALMLPLIGLRR398 pKa = 11.84ASDD401 pKa = 3.29QNEE404 pKa = 3.14IRR406 pKa = 11.84RR407 pKa = 11.84RR408 pKa = 11.84IQSRR412 pKa = 11.84GKK414 pKa = 10.16HH415 pKa = 4.78NLIPAGDD422 pKa = 3.8VVDD425 pKa = 4.27RR426 pKa = 11.84STWVQSLVSGTQVEE440 pKa = 4.74EE441 pKa = 4.17EE442 pKa = 4.16
MM1 pKa = 7.54AVGLGWRR8 pKa = 11.84VIACNRR14 pKa = 11.84LRR16 pKa = 11.84YY17 pKa = 9.65LCLLNFCLTRR27 pKa = 11.84NTNNKK32 pKa = 9.61RR33 pKa = 11.84NTNRR37 pKa = 11.84SQRR40 pKa = 11.84VAAPVATGLRR50 pKa = 11.84TSGGRR55 pKa = 11.84PQTTPAGAGAIRR67 pKa = 11.84VRR69 pKa = 11.84HH70 pKa = 5.85RR71 pKa = 11.84EE72 pKa = 3.84FIKK75 pKa = 10.98NITSEE80 pKa = 4.14HH81 pKa = 5.28TFEE84 pKa = 5.38NGVLEE89 pKa = 4.29LGINAGDD96 pKa = 3.45TEE98 pKa = 4.34MFPWLSNIANGYY110 pKa = 8.18EE111 pKa = 4.15RR112 pKa = 11.84YY113 pKa = 9.4CVNSMSISYY122 pKa = 10.14EE123 pKa = 3.86PFVSTTEE130 pKa = 3.83SGAVIMQVDD139 pKa = 4.58YY140 pKa = 11.38DD141 pKa = 4.22PADD144 pKa = 3.8EE145 pKa = 4.55PPISKK150 pKa = 9.74SHH152 pKa = 5.77MLNSLGATRR161 pKa = 11.84SAVWMRR167 pKa = 11.84SAMALNRR174 pKa = 11.84KK175 pKa = 7.73EE176 pKa = 4.86LSYY179 pKa = 11.33DD180 pKa = 2.93DD181 pKa = 4.41HH182 pKa = 8.57LFVRR186 pKa = 11.84HH187 pKa = 5.7AVRR190 pKa = 11.84AGFSEE195 pKa = 3.79NLKK198 pKa = 10.56LYY200 pKa = 10.67DD201 pKa = 3.43VGTVFIALTDD211 pKa = 3.77VPAPTTLSYY220 pKa = 11.58GEE222 pKa = 4.06VWVTYY227 pKa = 10.08DD228 pKa = 2.94ITLMVPAFHH237 pKa = 6.62KK238 pKa = 10.59QEE240 pKa = 4.23PNAAEE245 pKa = 4.22SKK247 pKa = 10.45VNQSNYY253 pKa = 9.85FDD255 pKa = 3.49ILGTISSDD263 pKa = 3.51RR264 pKa = 11.84PNTMLQGSSINFTTDD279 pKa = 2.37SNNAGEE285 pKa = 4.27CAITFQEE292 pKa = 4.68PFTGLLYY299 pKa = 10.86LEE301 pKa = 4.14QSGYY305 pKa = 11.31AEE307 pKa = 4.82DD308 pKa = 5.04SITHH312 pKa = 6.75LEE314 pKa = 4.32LEE316 pKa = 4.62PTTTPADD323 pKa = 3.13NWITKK328 pKa = 9.03LARR331 pKa = 11.84LGGGVIDD338 pKa = 6.63YY339 pKa = 10.91ILLDD343 pKa = 4.05NKK345 pKa = 9.24WKK347 pKa = 10.59YY348 pKa = 10.56LIEE351 pKa = 4.22VVADD355 pKa = 3.7AGEE358 pKa = 4.56SIVFDD363 pKa = 4.19ALAVGAGDD371 pKa = 3.57LTTWIGEE378 pKa = 4.16TAMALSPYY386 pKa = 10.46AEE388 pKa = 4.22ALMLPLIGLRR398 pKa = 11.84ASDD401 pKa = 3.29QNEE404 pKa = 3.14IRR406 pKa = 11.84RR407 pKa = 11.84RR408 pKa = 11.84IQSRR412 pKa = 11.84GKK414 pKa = 10.16HH415 pKa = 4.78NLIPAGDD422 pKa = 3.8VVDD425 pKa = 4.27RR426 pKa = 11.84STWVQSLVSGTQVEE440 pKa = 4.74EE441 pKa = 4.17EE442 pKa = 4.16
Molecular weight: 48.72 kDa
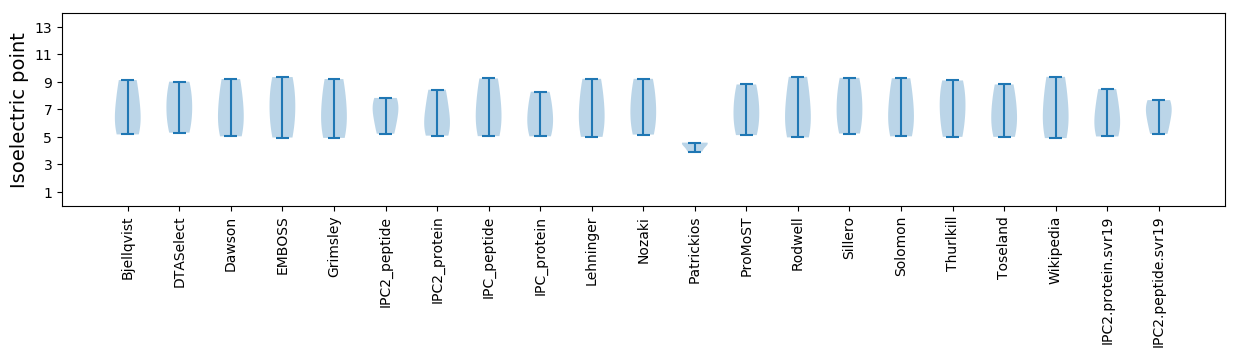
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGZ6|A0A1L3KGZ6_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 4 OX=1922725 PE=4 SV=1
MM1 pKa = 7.62EE2 pKa = 5.23KK3 pKa = 10.74GEE5 pKa = 4.41RR6 pKa = 11.84IRR8 pKa = 11.84IRR10 pKa = 11.84KK11 pKa = 8.06ARR13 pKa = 11.84QVGRR17 pKa = 11.84SKK19 pKa = 10.79KK20 pKa = 9.32ARR22 pKa = 11.84TYY24 pKa = 10.52YY25 pKa = 10.15RR26 pKa = 11.84VNGGNGPDD34 pKa = 3.15WDD36 pKa = 4.22IPNNDD41 pKa = 3.49IEE43 pKa = 4.89TVTHH47 pKa = 6.71AVLEE51 pKa = 4.32RR52 pKa = 11.84VFFVKK57 pKa = 10.34DD58 pKa = 3.43GKK60 pKa = 11.21GGFQRR65 pKa = 11.84APKK68 pKa = 8.48PWSHH72 pKa = 6.97SSINAQEE79 pKa = 4.22NPVAQARR86 pKa = 11.84SYY88 pKa = 11.0VQDD91 pKa = 3.59KK92 pKa = 10.64LSQFSRR98 pKa = 11.84EE99 pKa = 3.87MEE101 pKa = 4.19RR102 pKa = 11.84CSKK105 pKa = 10.01IHH107 pKa = 6.26GVVSPCTNQEE117 pKa = 3.76FVEE120 pKa = 5.06CYY122 pKa = 10.25GGAKK126 pKa = 9.75RR127 pKa = 11.84KK128 pKa = 9.64IYY130 pKa = 9.12EE131 pKa = 3.94QAVEE135 pKa = 4.18SLEE138 pKa = 4.36SNPLQARR145 pKa = 11.84DD146 pKa = 3.42KK147 pKa = 10.21RR148 pKa = 11.84VKK150 pKa = 10.62VFTKK154 pKa = 10.62DD155 pKa = 3.11EE156 pKa = 4.04YY157 pKa = 11.13LKK159 pKa = 10.76PDD161 pKa = 3.92GAPRR165 pKa = 11.84AIQPRR170 pKa = 11.84SPRR173 pKa = 11.84FNVCLGRR180 pKa = 11.84YY181 pKa = 7.97VKK183 pKa = 10.37QLEE186 pKa = 4.17HH187 pKa = 7.31LIFEE191 pKa = 5.59AINEE195 pKa = 4.08IFDD198 pKa = 3.59GSGEE202 pKa = 4.28HH203 pKa = 5.5KK204 pKa = 9.63TVAKK208 pKa = 9.35GMNMNEE214 pKa = 3.89RR215 pKa = 11.84GEE217 pKa = 4.49EE218 pKa = 3.7IRR220 pKa = 11.84GMWEE224 pKa = 3.8KK225 pKa = 10.75YY226 pKa = 8.76VNPVAVGLDD235 pKa = 3.14ASRR238 pKa = 11.84FDD240 pKa = 3.43QHH242 pKa = 7.83INTLLLEE249 pKa = 4.72HH250 pKa = 6.09EE251 pKa = 4.35HH252 pKa = 6.66KK253 pKa = 10.26IYY255 pKa = 10.94RR256 pKa = 11.84MWSTGQGDD264 pKa = 4.77DD265 pKa = 4.8LPNLNTLLSAQLKK278 pKa = 9.67NKK280 pKa = 9.23GVYY283 pKa = 9.97VGIDD287 pKa = 3.82GILKK291 pKa = 10.31YY292 pKa = 10.29KK293 pKa = 11.01VNGCRR298 pKa = 11.84MSGDD302 pKa = 3.63MNTSLGNVIIMCSLMYY318 pKa = 10.51SYY320 pKa = 11.06FKK322 pKa = 10.57DD323 pKa = 3.35KK324 pKa = 11.57NMLGKK329 pKa = 10.14ISLLNDD335 pKa = 3.39GDD337 pKa = 4.0DD338 pKa = 3.94CVIIMEE344 pKa = 4.2QKK346 pKa = 9.92RR347 pKa = 11.84LKK349 pKa = 10.7EE350 pKa = 3.97FTRR353 pKa = 11.84GLKK356 pKa = 10.11DD357 pKa = 2.67WFLRR361 pKa = 11.84MGITMEE367 pKa = 3.88FDD369 pKa = 3.21GVYY372 pKa = 8.94NTLEE376 pKa = 3.96EE377 pKa = 4.85VEE379 pKa = 4.66FCQARR384 pKa = 11.84PVFNEE389 pKa = 3.31EE390 pKa = 3.58LGYY393 pKa = 11.21VLTPRR398 pKa = 11.84PSKK401 pKa = 10.46RR402 pKa = 11.84LYY404 pKa = 10.86SDD406 pKa = 4.16VISTKK411 pKa = 7.74MMKK414 pKa = 10.0SKK416 pKa = 10.5KK417 pKa = 9.85VYY419 pKa = 9.88RR420 pKa = 11.84KK421 pKa = 9.51QLGAIAGCGLAMSGGVPIFQQFYY444 pKa = 7.54TWMGRR449 pKa = 11.84GATPWVPKK457 pKa = 10.37MGDD460 pKa = 2.68HH461 pKa = 6.43YY462 pKa = 11.54YY463 pKa = 11.06KK464 pKa = 10.54FRR466 pKa = 11.84QEE468 pKa = 5.26LIDD471 pKa = 3.81GMTYY475 pKa = 10.07KK476 pKa = 10.32QRR478 pKa = 11.84EE479 pKa = 3.82PTMRR483 pKa = 11.84EE484 pKa = 3.49RR485 pKa = 11.84ISFYY489 pKa = 9.55FAHH492 pKa = 7.46GITPAEE498 pKa = 3.95QKK500 pKa = 10.81LIEE503 pKa = 4.82NYY505 pKa = 8.48YY506 pKa = 10.36QCLPDD511 pKa = 4.64PLWSKK516 pKa = 10.34PVIDD520 pKa = 4.02PEE522 pKa = 4.29RR523 pKa = 11.84YY524 pKa = 8.52IDD526 pKa = 3.64SVQYY530 pKa = 10.67LVEE533 pKa = 4.34PEE535 pKa = 4.06QKK537 pKa = 10.08FKK539 pKa = 11.07RR540 pKa = 11.84LSS542 pKa = 3.27
MM1 pKa = 7.62EE2 pKa = 5.23KK3 pKa = 10.74GEE5 pKa = 4.41RR6 pKa = 11.84IRR8 pKa = 11.84IRR10 pKa = 11.84KK11 pKa = 8.06ARR13 pKa = 11.84QVGRR17 pKa = 11.84SKK19 pKa = 10.79KK20 pKa = 9.32ARR22 pKa = 11.84TYY24 pKa = 10.52YY25 pKa = 10.15RR26 pKa = 11.84VNGGNGPDD34 pKa = 3.15WDD36 pKa = 4.22IPNNDD41 pKa = 3.49IEE43 pKa = 4.89TVTHH47 pKa = 6.71AVLEE51 pKa = 4.32RR52 pKa = 11.84VFFVKK57 pKa = 10.34DD58 pKa = 3.43GKK60 pKa = 11.21GGFQRR65 pKa = 11.84APKK68 pKa = 8.48PWSHH72 pKa = 6.97SSINAQEE79 pKa = 4.22NPVAQARR86 pKa = 11.84SYY88 pKa = 11.0VQDD91 pKa = 3.59KK92 pKa = 10.64LSQFSRR98 pKa = 11.84EE99 pKa = 3.87MEE101 pKa = 4.19RR102 pKa = 11.84CSKK105 pKa = 10.01IHH107 pKa = 6.26GVVSPCTNQEE117 pKa = 3.76FVEE120 pKa = 5.06CYY122 pKa = 10.25GGAKK126 pKa = 9.75RR127 pKa = 11.84KK128 pKa = 9.64IYY130 pKa = 9.12EE131 pKa = 3.94QAVEE135 pKa = 4.18SLEE138 pKa = 4.36SNPLQARR145 pKa = 11.84DD146 pKa = 3.42KK147 pKa = 10.21RR148 pKa = 11.84VKK150 pKa = 10.62VFTKK154 pKa = 10.62DD155 pKa = 3.11EE156 pKa = 4.04YY157 pKa = 11.13LKK159 pKa = 10.76PDD161 pKa = 3.92GAPRR165 pKa = 11.84AIQPRR170 pKa = 11.84SPRR173 pKa = 11.84FNVCLGRR180 pKa = 11.84YY181 pKa = 7.97VKK183 pKa = 10.37QLEE186 pKa = 4.17HH187 pKa = 7.31LIFEE191 pKa = 5.59AINEE195 pKa = 4.08IFDD198 pKa = 3.59GSGEE202 pKa = 4.28HH203 pKa = 5.5KK204 pKa = 9.63TVAKK208 pKa = 9.35GMNMNEE214 pKa = 3.89RR215 pKa = 11.84GEE217 pKa = 4.49EE218 pKa = 3.7IRR220 pKa = 11.84GMWEE224 pKa = 3.8KK225 pKa = 10.75YY226 pKa = 8.76VNPVAVGLDD235 pKa = 3.14ASRR238 pKa = 11.84FDD240 pKa = 3.43QHH242 pKa = 7.83INTLLLEE249 pKa = 4.72HH250 pKa = 6.09EE251 pKa = 4.35HH252 pKa = 6.66KK253 pKa = 10.26IYY255 pKa = 10.94RR256 pKa = 11.84MWSTGQGDD264 pKa = 4.77DD265 pKa = 4.8LPNLNTLLSAQLKK278 pKa = 9.67NKK280 pKa = 9.23GVYY283 pKa = 9.97VGIDD287 pKa = 3.82GILKK291 pKa = 10.31YY292 pKa = 10.29KK293 pKa = 11.01VNGCRR298 pKa = 11.84MSGDD302 pKa = 3.63MNTSLGNVIIMCSLMYY318 pKa = 10.51SYY320 pKa = 11.06FKK322 pKa = 10.57DD323 pKa = 3.35KK324 pKa = 11.57NMLGKK329 pKa = 10.14ISLLNDD335 pKa = 3.39GDD337 pKa = 4.0DD338 pKa = 3.94CVIIMEE344 pKa = 4.2QKK346 pKa = 9.92RR347 pKa = 11.84LKK349 pKa = 10.7EE350 pKa = 3.97FTRR353 pKa = 11.84GLKK356 pKa = 10.11DD357 pKa = 2.67WFLRR361 pKa = 11.84MGITMEE367 pKa = 3.88FDD369 pKa = 3.21GVYY372 pKa = 8.94NTLEE376 pKa = 3.96EE377 pKa = 4.85VEE379 pKa = 4.66FCQARR384 pKa = 11.84PVFNEE389 pKa = 3.31EE390 pKa = 3.58LGYY393 pKa = 11.21VLTPRR398 pKa = 11.84PSKK401 pKa = 10.46RR402 pKa = 11.84LYY404 pKa = 10.86SDD406 pKa = 4.16VISTKK411 pKa = 7.74MMKK414 pKa = 10.0SKK416 pKa = 10.5KK417 pKa = 9.85VYY419 pKa = 9.88RR420 pKa = 11.84KK421 pKa = 9.51QLGAIAGCGLAMSGGVPIFQQFYY444 pKa = 7.54TWMGRR449 pKa = 11.84GATPWVPKK457 pKa = 10.37MGDD460 pKa = 2.68HH461 pKa = 6.43YY462 pKa = 11.54YY463 pKa = 11.06KK464 pKa = 10.54FRR466 pKa = 11.84QEE468 pKa = 5.26LIDD471 pKa = 3.81GMTYY475 pKa = 10.07KK476 pKa = 10.32QRR478 pKa = 11.84EE479 pKa = 3.82PTMRR483 pKa = 11.84EE484 pKa = 3.49RR485 pKa = 11.84ISFYY489 pKa = 9.55FAHH492 pKa = 7.46GITPAEE498 pKa = 3.95QKK500 pKa = 10.81LIEE503 pKa = 4.82NYY505 pKa = 8.48YY506 pKa = 10.36QCLPDD511 pKa = 4.64PLWSKK516 pKa = 10.34PVIDD520 pKa = 4.02PEE522 pKa = 4.29RR523 pKa = 11.84YY524 pKa = 8.52IDD526 pKa = 3.64SVQYY530 pKa = 10.67LVEE533 pKa = 4.34PEE535 pKa = 4.06QKK537 pKa = 10.08FKK539 pKa = 11.07RR540 pKa = 11.84LSS542 pKa = 3.27
Molecular weight: 62.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1355 |
371 |
542 |
451.7 |
50.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.528 ± 1.602 | 1.402 ± 0.221 |
4.945 ± 0.032 | 7.306 ± 0.447 |
3.173 ± 0.523 | 8.044 ± 0.378 |
1.845 ± 0.015 | 5.387 ± 0.356 |
5.166 ± 1.386 | 7.749 ± 0.542 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.247 ± 0.33 | 5.166 ± 0.348 |
4.502 ± 0.146 | 3.321 ± 0.66 |
7.159 ± 0.588 | 6.347 ± 0.451 |
5.461 ± 0.986 | 6.863 ± 0.151 |
1.624 ± 0.079 | 3.764 ± 0.542 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
