
CrAssphage sp.
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae; crAss-like viruses
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
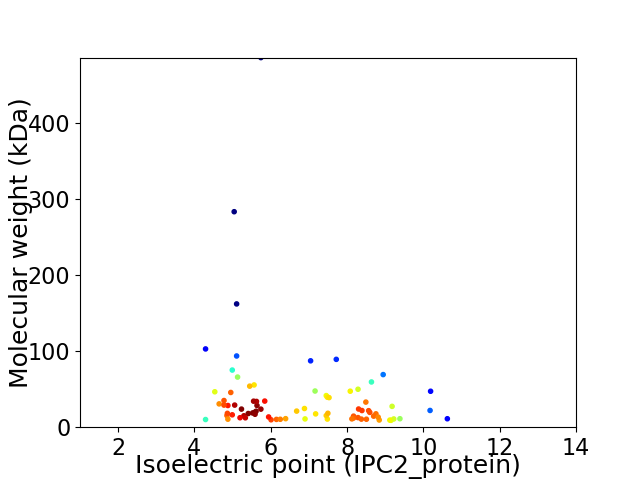
Virtual 2D-PAGE plot for 80 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
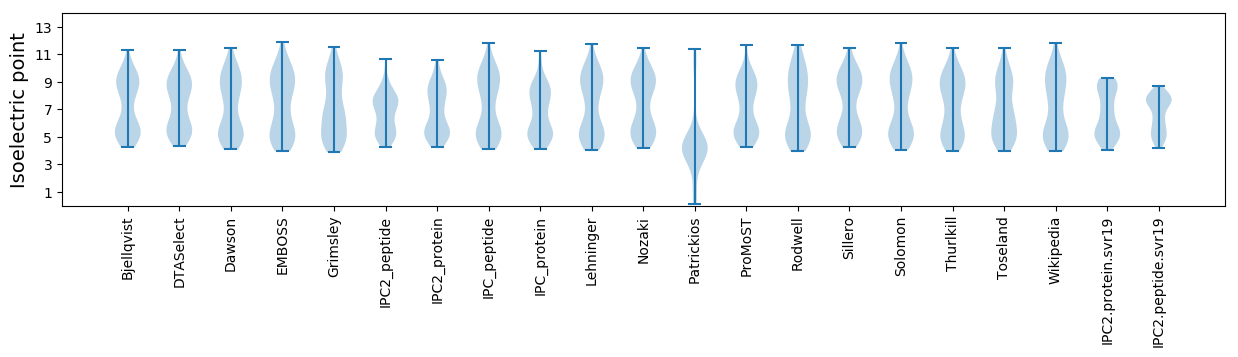
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345BP28|A0A345BP28_9CAUD Putative tail tubular protein OS=CrAssphage sp. OX=2212563 PE=4 SV=1
MM1 pKa = 7.64KK2 pKa = 10.25LYY4 pKa = 10.53KK5 pKa = 10.66DD6 pKa = 3.73NLGKK10 pKa = 10.53VAITVEE16 pKa = 3.64QGYY19 pKa = 9.4WDD21 pKa = 3.77EE22 pKa = 4.54NKK24 pKa = 10.7DD25 pKa = 3.66YY26 pKa = 11.56DD27 pKa = 3.87KK28 pKa = 10.87LTVVEE33 pKa = 4.3RR34 pKa = 11.84EE35 pKa = 4.69GIFGTYY41 pKa = 9.35ISRR44 pKa = 11.84KK45 pKa = 7.74PVPAGTEE52 pKa = 3.95LSDD55 pKa = 3.18RR56 pKa = 11.84TYY58 pKa = 10.2WIPFSSLKK66 pKa = 10.52EE67 pKa = 3.95EE68 pKa = 3.87IVINYY73 pKa = 9.97NEE75 pKa = 4.91FISHH79 pKa = 6.7YY80 pKa = 10.93GPMLDD85 pKa = 5.08DD86 pKa = 3.47QAEE89 pKa = 4.31VLNNVQLLINEE100 pKa = 5.03IYY102 pKa = 10.72NKK104 pKa = 9.96LPSGIIFEE112 pKa = 4.34GDD114 pKa = 2.92KK115 pKa = 10.72TFIRR119 pKa = 11.84PGHH122 pKa = 6.12PEE124 pKa = 4.42EE125 pKa = 4.09ITLTGSTVNEE135 pKa = 4.22DD136 pKa = 3.21PATFKK141 pKa = 10.83LYY143 pKa = 10.76KK144 pKa = 9.87NSKK147 pKa = 9.35LVHH150 pKa = 5.35TVTAKK155 pKa = 10.48SFTYY159 pKa = 10.21KK160 pKa = 10.72AIADD164 pKa = 3.98DD165 pKa = 3.64EE166 pKa = 4.55CAYY169 pKa = 10.17KK170 pKa = 10.75LIVEE174 pKa = 4.2QSGYY178 pKa = 10.61RR179 pKa = 11.84YY180 pKa = 9.31VAGWNITLRR189 pKa = 11.84YY190 pKa = 8.34PIYY193 pKa = 10.17AGSGATYY200 pKa = 9.97EE201 pKa = 5.06DD202 pKa = 4.06VVSDD206 pKa = 3.68YY207 pKa = 11.53NIIKK211 pKa = 9.97IDD213 pKa = 3.69TNPEE217 pKa = 3.12GLYY220 pKa = 10.11IINVINDD227 pKa = 3.03GDD229 pKa = 4.35YY230 pKa = 11.14IFFVAPKK237 pKa = 9.83NVIIHH242 pKa = 5.95SIKK245 pKa = 10.89LNGLDD250 pKa = 3.56VPLKK254 pKa = 8.6QTEE257 pKa = 4.22EE258 pKa = 4.31VEE260 pKa = 4.8LEE262 pKa = 4.01TEE264 pKa = 4.24YY265 pKa = 10.48IIYY268 pKa = 10.59QSDD271 pKa = 3.19NTYY274 pKa = 10.89EE275 pKa = 4.22SNNYY279 pKa = 8.85PIYY282 pKa = 9.56INSYY286 pKa = 8.8EE287 pKa = 4.09GLNGGDD293 pKa = 3.49FVKK296 pKa = 10.59ILDD299 pKa = 3.75VLQDD303 pKa = 3.52KK304 pKa = 8.12VTSEE308 pKa = 4.85DD309 pKa = 2.65ITRR312 pKa = 11.84ITNEE316 pKa = 3.68NLEE319 pKa = 4.29IKK321 pKa = 8.9QTLQQVKK328 pKa = 10.29EE329 pKa = 4.25EE330 pKa = 4.13NNEE333 pKa = 3.75VNNNIDD339 pKa = 3.21AALSVVEE346 pKa = 4.55TYY348 pKa = 8.69KK349 pKa = 10.69TEE351 pKa = 4.12INKK354 pKa = 9.98SLAIIEE360 pKa = 4.7NISNEE365 pKa = 4.0LNDD368 pKa = 3.02ITEE371 pKa = 5.01NINTSNVDD379 pKa = 3.83LNVLKK384 pKa = 10.37TEE386 pKa = 4.21VTNMASSVNSLNKK399 pKa = 10.21SLEE402 pKa = 4.03EE403 pKa = 4.33LGTNVYY409 pKa = 10.43SAIDD413 pKa = 3.53AVQEE417 pKa = 3.96NVDD420 pKa = 4.08RR421 pKa = 11.84IGEE424 pKa = 4.13GRR426 pKa = 11.84VDD428 pKa = 4.72IINEE432 pKa = 3.83DD433 pKa = 3.52GFRR436 pKa = 11.84NNNVIYY442 pKa = 10.2LKK444 pKa = 10.65PNWTKK449 pKa = 10.34VSVYY453 pKa = 10.34GNEE456 pKa = 3.97PEE458 pKa = 4.08IVPVLTQNMINRR470 pKa = 11.84PYY472 pKa = 7.32TTYY475 pKa = 10.95VIDD478 pKa = 3.47YY479 pKa = 9.41NYY481 pKa = 10.92YY482 pKa = 9.52IFDD485 pKa = 4.49TIEE488 pKa = 4.13LPIGTTLKK496 pKa = 10.36FRR498 pKa = 11.84AGALLFVEE506 pKa = 4.53NGSMYY511 pKa = 10.57SQYY514 pKa = 11.51NCIIDD519 pKa = 4.1DD520 pKa = 4.27CGNNNIIQGEE530 pKa = 4.37TLAMQIVFSNVYY542 pKa = 10.46LDD544 pKa = 3.44WFYY547 pKa = 11.81LFLDD551 pKa = 3.25IGGSYY556 pKa = 10.39VEE558 pKa = 4.48NLNGILDD565 pKa = 3.93YY566 pKa = 11.11AFSIARR572 pKa = 11.84GPIRR576 pKa = 11.84FRR578 pKa = 11.84QVDD581 pKa = 3.73RR582 pKa = 11.84LVIDD586 pKa = 4.29GFVNACVGTIDD597 pKa = 5.54AEE599 pKa = 4.52SKK601 pKa = 10.71SININNCEE609 pKa = 3.93NATITIQNSNSVIIEE624 pKa = 4.01KK625 pKa = 10.39ADD627 pKa = 4.01NISDD631 pKa = 3.61VSEE634 pKa = 3.94ICIKK638 pKa = 10.9NSIIDD643 pKa = 3.77VINIYY648 pKa = 9.89SPCTLSLRR656 pKa = 11.84IDD658 pKa = 3.35NCNINEE664 pKa = 4.02IQAVCDD670 pKa = 4.25DD671 pKa = 3.93GVLTIEE677 pKa = 5.16NSIIQNNIVSSFYY690 pKa = 11.19TNIIRR695 pKa = 11.84NNVMLHH701 pKa = 4.93TAYY704 pKa = 9.8ISFVQSGIYY713 pKa = 9.72KK714 pKa = 9.96ILEE717 pKa = 4.15APKK720 pKa = 9.57TAIIDD725 pKa = 3.55NNIIGEE731 pKa = 4.34VILDD735 pKa = 3.65EE736 pKa = 6.15FEE738 pKa = 4.58FLDD741 pKa = 5.06DD742 pKa = 3.8FTTAIPIIFEE752 pKa = 4.04FYY754 pKa = 10.76GYY756 pKa = 9.72IPHH759 pKa = 6.69YY760 pKa = 9.87NFSFIINNTNVDD772 pKa = 4.38LNIQNDD778 pKa = 3.44NKK780 pKa = 10.47FAGIISNSNVYY791 pKa = 10.44LHH793 pKa = 6.54LHH795 pKa = 5.66SLNTALEE802 pKa = 4.44TYY804 pKa = 10.52EE805 pKa = 4.7NVLLYY810 pKa = 8.88EE811 pKa = 4.56TFLHH815 pKa = 7.22LYY817 pKa = 8.89NCVVEE822 pKa = 4.33NSPNSEE828 pKa = 3.9VSSSDD833 pKa = 3.37DD834 pKa = 3.34YY835 pKa = 11.78CIISSTLTFDD845 pKa = 2.92NCYY848 pKa = 10.87ANNLGTIVGYY858 pKa = 5.9EE859 pKa = 3.69TSYY862 pKa = 11.05IRR864 pKa = 11.84TNNSPISVNEE874 pKa = 3.8DD875 pKa = 2.94EE876 pKa = 5.29LLYY879 pKa = 11.08GHH881 pKa = 7.24TYY883 pKa = 10.34DD884 pKa = 3.53ISRR887 pKa = 11.84GTVINTEE894 pKa = 3.92NKK896 pKa = 10.19LGISRR901 pKa = 11.84GRR903 pKa = 11.84NKK905 pKa = 10.45EE906 pKa = 4.05PLWLEE911 pKa = 3.92
MM1 pKa = 7.64KK2 pKa = 10.25LYY4 pKa = 10.53KK5 pKa = 10.66DD6 pKa = 3.73NLGKK10 pKa = 10.53VAITVEE16 pKa = 3.64QGYY19 pKa = 9.4WDD21 pKa = 3.77EE22 pKa = 4.54NKK24 pKa = 10.7DD25 pKa = 3.66YY26 pKa = 11.56DD27 pKa = 3.87KK28 pKa = 10.87LTVVEE33 pKa = 4.3RR34 pKa = 11.84EE35 pKa = 4.69GIFGTYY41 pKa = 9.35ISRR44 pKa = 11.84KK45 pKa = 7.74PVPAGTEE52 pKa = 3.95LSDD55 pKa = 3.18RR56 pKa = 11.84TYY58 pKa = 10.2WIPFSSLKK66 pKa = 10.52EE67 pKa = 3.95EE68 pKa = 3.87IVINYY73 pKa = 9.97NEE75 pKa = 4.91FISHH79 pKa = 6.7YY80 pKa = 10.93GPMLDD85 pKa = 5.08DD86 pKa = 3.47QAEE89 pKa = 4.31VLNNVQLLINEE100 pKa = 5.03IYY102 pKa = 10.72NKK104 pKa = 9.96LPSGIIFEE112 pKa = 4.34GDD114 pKa = 2.92KK115 pKa = 10.72TFIRR119 pKa = 11.84PGHH122 pKa = 6.12PEE124 pKa = 4.42EE125 pKa = 4.09ITLTGSTVNEE135 pKa = 4.22DD136 pKa = 3.21PATFKK141 pKa = 10.83LYY143 pKa = 10.76KK144 pKa = 9.87NSKK147 pKa = 9.35LVHH150 pKa = 5.35TVTAKK155 pKa = 10.48SFTYY159 pKa = 10.21KK160 pKa = 10.72AIADD164 pKa = 3.98DD165 pKa = 3.64EE166 pKa = 4.55CAYY169 pKa = 10.17KK170 pKa = 10.75LIVEE174 pKa = 4.2QSGYY178 pKa = 10.61RR179 pKa = 11.84YY180 pKa = 9.31VAGWNITLRR189 pKa = 11.84YY190 pKa = 8.34PIYY193 pKa = 10.17AGSGATYY200 pKa = 9.97EE201 pKa = 5.06DD202 pKa = 4.06VVSDD206 pKa = 3.68YY207 pKa = 11.53NIIKK211 pKa = 9.97IDD213 pKa = 3.69TNPEE217 pKa = 3.12GLYY220 pKa = 10.11IINVINDD227 pKa = 3.03GDD229 pKa = 4.35YY230 pKa = 11.14IFFVAPKK237 pKa = 9.83NVIIHH242 pKa = 5.95SIKK245 pKa = 10.89LNGLDD250 pKa = 3.56VPLKK254 pKa = 8.6QTEE257 pKa = 4.22EE258 pKa = 4.31VEE260 pKa = 4.8LEE262 pKa = 4.01TEE264 pKa = 4.24YY265 pKa = 10.48IIYY268 pKa = 10.59QSDD271 pKa = 3.19NTYY274 pKa = 10.89EE275 pKa = 4.22SNNYY279 pKa = 8.85PIYY282 pKa = 9.56INSYY286 pKa = 8.8EE287 pKa = 4.09GLNGGDD293 pKa = 3.49FVKK296 pKa = 10.59ILDD299 pKa = 3.75VLQDD303 pKa = 3.52KK304 pKa = 8.12VTSEE308 pKa = 4.85DD309 pKa = 2.65ITRR312 pKa = 11.84ITNEE316 pKa = 3.68NLEE319 pKa = 4.29IKK321 pKa = 8.9QTLQQVKK328 pKa = 10.29EE329 pKa = 4.25EE330 pKa = 4.13NNEE333 pKa = 3.75VNNNIDD339 pKa = 3.21AALSVVEE346 pKa = 4.55TYY348 pKa = 8.69KK349 pKa = 10.69TEE351 pKa = 4.12INKK354 pKa = 9.98SLAIIEE360 pKa = 4.7NISNEE365 pKa = 4.0LNDD368 pKa = 3.02ITEE371 pKa = 5.01NINTSNVDD379 pKa = 3.83LNVLKK384 pKa = 10.37TEE386 pKa = 4.21VTNMASSVNSLNKK399 pKa = 10.21SLEE402 pKa = 4.03EE403 pKa = 4.33LGTNVYY409 pKa = 10.43SAIDD413 pKa = 3.53AVQEE417 pKa = 3.96NVDD420 pKa = 4.08RR421 pKa = 11.84IGEE424 pKa = 4.13GRR426 pKa = 11.84VDD428 pKa = 4.72IINEE432 pKa = 3.83DD433 pKa = 3.52GFRR436 pKa = 11.84NNNVIYY442 pKa = 10.2LKK444 pKa = 10.65PNWTKK449 pKa = 10.34VSVYY453 pKa = 10.34GNEE456 pKa = 3.97PEE458 pKa = 4.08IVPVLTQNMINRR470 pKa = 11.84PYY472 pKa = 7.32TTYY475 pKa = 10.95VIDD478 pKa = 3.47YY479 pKa = 9.41NYY481 pKa = 10.92YY482 pKa = 9.52IFDD485 pKa = 4.49TIEE488 pKa = 4.13LPIGTTLKK496 pKa = 10.36FRR498 pKa = 11.84AGALLFVEE506 pKa = 4.53NGSMYY511 pKa = 10.57SQYY514 pKa = 11.51NCIIDD519 pKa = 4.1DD520 pKa = 4.27CGNNNIIQGEE530 pKa = 4.37TLAMQIVFSNVYY542 pKa = 10.46LDD544 pKa = 3.44WFYY547 pKa = 11.81LFLDD551 pKa = 3.25IGGSYY556 pKa = 10.39VEE558 pKa = 4.48NLNGILDD565 pKa = 3.93YY566 pKa = 11.11AFSIARR572 pKa = 11.84GPIRR576 pKa = 11.84FRR578 pKa = 11.84QVDD581 pKa = 3.73RR582 pKa = 11.84LVIDD586 pKa = 4.29GFVNACVGTIDD597 pKa = 5.54AEE599 pKa = 4.52SKK601 pKa = 10.71SININNCEE609 pKa = 3.93NATITIQNSNSVIIEE624 pKa = 4.01KK625 pKa = 10.39ADD627 pKa = 4.01NISDD631 pKa = 3.61VSEE634 pKa = 3.94ICIKK638 pKa = 10.9NSIIDD643 pKa = 3.77VINIYY648 pKa = 9.89SPCTLSLRR656 pKa = 11.84IDD658 pKa = 3.35NCNINEE664 pKa = 4.02IQAVCDD670 pKa = 4.25DD671 pKa = 3.93GVLTIEE677 pKa = 5.16NSIIQNNIVSSFYY690 pKa = 11.19TNIIRR695 pKa = 11.84NNVMLHH701 pKa = 4.93TAYY704 pKa = 9.8ISFVQSGIYY713 pKa = 9.72KK714 pKa = 9.96ILEE717 pKa = 4.15APKK720 pKa = 9.57TAIIDD725 pKa = 3.55NNIIGEE731 pKa = 4.34VILDD735 pKa = 3.65EE736 pKa = 6.15FEE738 pKa = 4.58FLDD741 pKa = 5.06DD742 pKa = 3.8FTTAIPIIFEE752 pKa = 4.04FYY754 pKa = 10.76GYY756 pKa = 9.72IPHH759 pKa = 6.69YY760 pKa = 9.87NFSFIINNTNVDD772 pKa = 4.38LNIQNDD778 pKa = 3.44NKK780 pKa = 10.47FAGIISNSNVYY791 pKa = 10.44LHH793 pKa = 6.54LHH795 pKa = 5.66SLNTALEE802 pKa = 4.44TYY804 pKa = 10.52EE805 pKa = 4.7NVLLYY810 pKa = 8.88EE811 pKa = 4.56TFLHH815 pKa = 7.22LYY817 pKa = 8.89NCVVEE822 pKa = 4.33NSPNSEE828 pKa = 3.9VSSSDD833 pKa = 3.37DD834 pKa = 3.34YY835 pKa = 11.78CIISSTLTFDD845 pKa = 2.92NCYY848 pKa = 10.87ANNLGTIVGYY858 pKa = 5.9EE859 pKa = 3.69TSYY862 pKa = 11.05IRR864 pKa = 11.84TNNSPISVNEE874 pKa = 3.8DD875 pKa = 2.94EE876 pKa = 5.29LLYY879 pKa = 11.08GHH881 pKa = 7.24TYY883 pKa = 10.34DD884 pKa = 3.53ISRR887 pKa = 11.84GTVINTEE894 pKa = 3.92NKK896 pKa = 10.19LGISRR901 pKa = 11.84GRR903 pKa = 11.84NKK905 pKa = 10.45EE906 pKa = 4.05PLWLEE911 pKa = 3.92
Molecular weight: 103.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345BNZ9|A0A345BNZ9_9CAUD Uncharacterized protein OS=CrAssphage sp. OX=2212563 PE=4 SV=1
MM1 pKa = 7.52NIAIRR6 pKa = 11.84YY7 pKa = 7.09KK8 pKa = 10.68FKK10 pKa = 10.75FNKK13 pKa = 8.15ITIFTICINLLINISISIYY32 pKa = 9.28YY33 pKa = 8.44LHH35 pKa = 7.25KK36 pKa = 10.54SIYY39 pKa = 7.95SKK41 pKa = 10.92KK42 pKa = 10.03IIRR45 pKa = 11.84STNMMTKK52 pKa = 9.91HH53 pKa = 6.03YY54 pKa = 10.04IASRR58 pKa = 11.84YY59 pKa = 9.58NKK61 pKa = 9.69FIFYY65 pKa = 10.39IKK67 pKa = 10.02ILYY70 pKa = 9.21KK71 pKa = 10.58CIISFSTFNKK81 pKa = 9.43NRR83 pKa = 11.84NNTYY87 pKa = 8.41TSIFIVSFSRR97 pKa = 11.84KK98 pKa = 8.17NNSRR102 pKa = 11.84KK103 pKa = 9.25FFSKK107 pKa = 10.5NSSNKK112 pKa = 9.39TIIRR116 pKa = 11.84SNIYY120 pKa = 9.22SRR122 pKa = 11.84CNISYY127 pKa = 10.65ILFCNLINRR136 pKa = 11.84CWKK139 pKa = 10.17INIKK143 pKa = 10.43FSVLTFYY150 pKa = 10.9IKK152 pKa = 9.86MYY154 pKa = 10.83HH155 pKa = 6.58NINLINNKK163 pKa = 9.77YY164 pKa = 9.97NILFIFIIILYY175 pKa = 9.69NILYY179 pKa = 9.61YY180 pKa = 10.1II181 pKa = 4.5
MM1 pKa = 7.52NIAIRR6 pKa = 11.84YY7 pKa = 7.09KK8 pKa = 10.68FKK10 pKa = 10.75FNKK13 pKa = 8.15ITIFTICINLLINISISIYY32 pKa = 9.28YY33 pKa = 8.44LHH35 pKa = 7.25KK36 pKa = 10.54SIYY39 pKa = 7.95SKK41 pKa = 10.92KK42 pKa = 10.03IIRR45 pKa = 11.84STNMMTKK52 pKa = 9.91HH53 pKa = 6.03YY54 pKa = 10.04IASRR58 pKa = 11.84YY59 pKa = 9.58NKK61 pKa = 9.69FIFYY65 pKa = 10.39IKK67 pKa = 10.02ILYY70 pKa = 9.21KK71 pKa = 10.58CIISFSTFNKK81 pKa = 9.43NRR83 pKa = 11.84NNTYY87 pKa = 8.41TSIFIVSFSRR97 pKa = 11.84KK98 pKa = 8.17NNSRR102 pKa = 11.84KK103 pKa = 9.25FFSKK107 pKa = 10.5NSSNKK112 pKa = 9.39TIIRR116 pKa = 11.84SNIYY120 pKa = 9.22SRR122 pKa = 11.84CNISYY127 pKa = 10.65ILFCNLINRR136 pKa = 11.84CWKK139 pKa = 10.17INIKK143 pKa = 10.43FSVLTFYY150 pKa = 10.9IKK152 pKa = 9.86MYY154 pKa = 10.83HH155 pKa = 6.58NINLINNKK163 pKa = 9.77YY164 pKa = 9.97NILFIFIIILYY175 pKa = 9.69NILYY179 pKa = 9.61YY180 pKa = 10.1II181 pKa = 4.5
Molecular weight: 22.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
27590 |
81 |
4253 |
344.9 |
39.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.625 ± 0.353 | 1.225 ± 0.185 |
6.292 ± 0.184 | 6.524 ± 0.296 |
4.215 ± 0.147 | 5.259 ± 0.26 |
1.591 ± 0.103 | 8.499 ± 0.418 |
8.307 ± 0.306 | 7.506 ± 0.176 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.222 ± 0.145 | 8.311 ± 0.26 |
2.588 ± 0.129 | 3.226 ± 0.241 |
3.987 ± 0.14 | 6.557 ± 0.198 |
6.444 ± 0.23 | 5.676 ± 0.205 |
0.819 ± 0.086 | 5.125 ± 0.251 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
