
Formosa sp. Hel1_31_208
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Formosa; unclassified Formosa
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
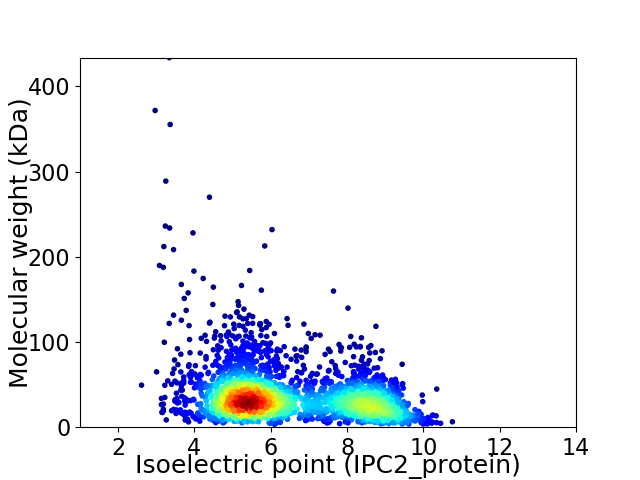
Virtual 2D-PAGE plot for 2771 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H1SG18|A0A1H1SG18_9FLAO Uncharacterized protein OS=Formosa sp. Hel1_31_208 OX=1798225 GN=SAMN04515667_2247 PE=4 SV=1
MM1 pKa = 7.32YY2 pKa = 10.28KK3 pKa = 10.66LSICSLLILLACFDD17 pKa = 4.16VLGQTTFTKK26 pKa = 8.92TQSIALPIPGGNAPYY41 pKa = 10.39NIASGLIDD49 pKa = 4.97DD50 pKa = 5.62DD51 pKa = 4.78SYY53 pKa = 11.99PDD55 pKa = 3.44IVVSTTLGDD64 pKa = 3.43TVYY67 pKa = 10.66WLRR70 pKa = 11.84NDD72 pKa = 3.24GTGNFTLQPTPIGILDD88 pKa = 3.68SAGGVAIADD97 pKa = 3.44INGNGFNDD105 pKa = 3.69VVSTSSIDD113 pKa = 3.42GTLVWFPNDD122 pKa = 3.04GLGNFGVEE130 pKa = 4.19QPISSSLNGPGQVYY144 pKa = 9.58VRR146 pKa = 11.84AIDD149 pKa = 3.57NNSTLDD155 pKa = 3.81VAVSAYY161 pKa = 10.4NGNEE165 pKa = 4.15VVWFANDD172 pKa = 3.35GLGNFSSKK180 pKa = 10.98NIINNTIPSPGAFAMKK196 pKa = 10.4DD197 pKa = 2.75IDD199 pKa = 3.98FDD201 pKa = 4.6GDD203 pKa = 3.32IDD205 pKa = 4.47SVIANAVAFGTPNDD219 pKa = 3.5CRR221 pKa = 11.84IEE223 pKa = 4.05VFYY226 pKa = 11.44NDD228 pKa = 3.28GDD230 pKa = 4.44GNFTADD236 pKa = 3.39TNAVSVNTKK245 pKa = 10.58DD246 pKa = 3.81YY247 pKa = 9.96IFSVMAEE254 pKa = 4.16DD255 pKa = 4.04VDD257 pKa = 4.1NDD259 pKa = 3.46SSLDD263 pKa = 3.51ILVTDD268 pKa = 4.16LTGNASWFKK277 pKa = 9.85RR278 pKa = 11.84TQISPGTATYY288 pKa = 10.53SEE290 pKa = 4.47TVIPSSIANPACLDD304 pKa = 4.82LRR306 pKa = 11.84DD307 pKa = 4.75LDD309 pKa = 4.08NDD311 pKa = 3.57NLKK314 pKa = 11.01DD315 pKa = 4.83FILTSAASGSGNDD328 pKa = 3.88IIWFKK333 pKa = 11.58GDD335 pKa = 3.01GLGNFGPEE343 pKa = 4.04QIIDD347 pKa = 3.49ATQNQAYY354 pKa = 7.69TITFADD360 pKa = 4.39FEE362 pKa = 4.89NDD364 pKa = 3.06GDD366 pKa = 5.12LDD368 pKa = 4.04TATIAYY374 pKa = 10.19ADD376 pKa = 3.6DD377 pKa = 3.49RR378 pKa = 11.84VNIFRR383 pKa = 11.84NEE385 pKa = 4.14NITLSLPAIDD395 pKa = 3.46SHH397 pKa = 5.73EE398 pKa = 4.27VKK400 pKa = 10.57VYY402 pKa = 9.6PNPVSNQLYY411 pKa = 8.74IQLNTEE417 pKa = 3.88VSFSISIYY425 pKa = 10.67NSIGQLIASQTISNNNFIDD444 pKa = 3.68VSHH447 pKa = 7.1FEE449 pKa = 4.09SGVYY453 pKa = 9.82FININNYY460 pKa = 7.85AKK462 pKa = 10.15SYY464 pKa = 10.97KK465 pKa = 9.08FVKK468 pKa = 10.12YY469 pKa = 10.57
MM1 pKa = 7.32YY2 pKa = 10.28KK3 pKa = 10.66LSICSLLILLACFDD17 pKa = 4.16VLGQTTFTKK26 pKa = 8.92TQSIALPIPGGNAPYY41 pKa = 10.39NIASGLIDD49 pKa = 4.97DD50 pKa = 5.62DD51 pKa = 4.78SYY53 pKa = 11.99PDD55 pKa = 3.44IVVSTTLGDD64 pKa = 3.43TVYY67 pKa = 10.66WLRR70 pKa = 11.84NDD72 pKa = 3.24GTGNFTLQPTPIGILDD88 pKa = 3.68SAGGVAIADD97 pKa = 3.44INGNGFNDD105 pKa = 3.69VVSTSSIDD113 pKa = 3.42GTLVWFPNDD122 pKa = 3.04GLGNFGVEE130 pKa = 4.19QPISSSLNGPGQVYY144 pKa = 9.58VRR146 pKa = 11.84AIDD149 pKa = 3.57NNSTLDD155 pKa = 3.81VAVSAYY161 pKa = 10.4NGNEE165 pKa = 4.15VVWFANDD172 pKa = 3.35GLGNFSSKK180 pKa = 10.98NIINNTIPSPGAFAMKK196 pKa = 10.4DD197 pKa = 2.75IDD199 pKa = 3.98FDD201 pKa = 4.6GDD203 pKa = 3.32IDD205 pKa = 4.47SVIANAVAFGTPNDD219 pKa = 3.5CRR221 pKa = 11.84IEE223 pKa = 4.05VFYY226 pKa = 11.44NDD228 pKa = 3.28GDD230 pKa = 4.44GNFTADD236 pKa = 3.39TNAVSVNTKK245 pKa = 10.58DD246 pKa = 3.81YY247 pKa = 9.96IFSVMAEE254 pKa = 4.16DD255 pKa = 4.04VDD257 pKa = 4.1NDD259 pKa = 3.46SSLDD263 pKa = 3.51ILVTDD268 pKa = 4.16LTGNASWFKK277 pKa = 9.85RR278 pKa = 11.84TQISPGTATYY288 pKa = 10.53SEE290 pKa = 4.47TVIPSSIANPACLDD304 pKa = 4.82LRR306 pKa = 11.84DD307 pKa = 4.75LDD309 pKa = 4.08NDD311 pKa = 3.57NLKK314 pKa = 11.01DD315 pKa = 4.83FILTSAASGSGNDD328 pKa = 3.88IIWFKK333 pKa = 11.58GDD335 pKa = 3.01GLGNFGPEE343 pKa = 4.04QIIDD347 pKa = 3.49ATQNQAYY354 pKa = 7.69TITFADD360 pKa = 4.39FEE362 pKa = 4.89NDD364 pKa = 3.06GDD366 pKa = 5.12LDD368 pKa = 4.04TATIAYY374 pKa = 10.19ADD376 pKa = 3.6DD377 pKa = 3.49RR378 pKa = 11.84VNIFRR383 pKa = 11.84NEE385 pKa = 4.14NITLSLPAIDD395 pKa = 3.46SHH397 pKa = 5.73EE398 pKa = 4.27VKK400 pKa = 10.57VYY402 pKa = 9.6PNPVSNQLYY411 pKa = 8.74IQLNTEE417 pKa = 3.88VSFSISIYY425 pKa = 10.67NSIGQLIASQTISNNNFIDD444 pKa = 3.68VSHH447 pKa = 7.1FEE449 pKa = 4.09SGVYY453 pKa = 9.82FININNYY460 pKa = 7.85AKK462 pKa = 10.15SYY464 pKa = 10.97KK465 pKa = 9.08FVKK468 pKa = 10.12YY469 pKa = 10.57
Molecular weight: 50.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H1U873|A0A1H1U873_9FLAO Colanic acid/amylovoran biosynthesis glycosyltransferase OS=Formosa sp. Hel1_31_208 OX=1798225 GN=SAMN04515667_2740 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
Molecular weight: 6.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
943956 |
29 |
4178 |
340.7 |
38.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.382 ± 0.045 | 0.796 ± 0.025 |
6.013 ± 0.057 | 6.295 ± 0.044 |
5.284 ± 0.042 | 6.271 ± 0.047 |
1.852 ± 0.027 | 8.135 ± 0.048 |
7.23 ± 0.085 | 9.181 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.232 ± 0.029 | 6.153 ± 0.051 |
3.321 ± 0.026 | 3.464 ± 0.028 |
3.414 ± 0.037 | 6.523 ± 0.038 |
6.173 ± 0.1 | 6.268 ± 0.036 |
1.022 ± 0.02 | 3.992 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
