
Pseudomonas phage phiPSA1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 52 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
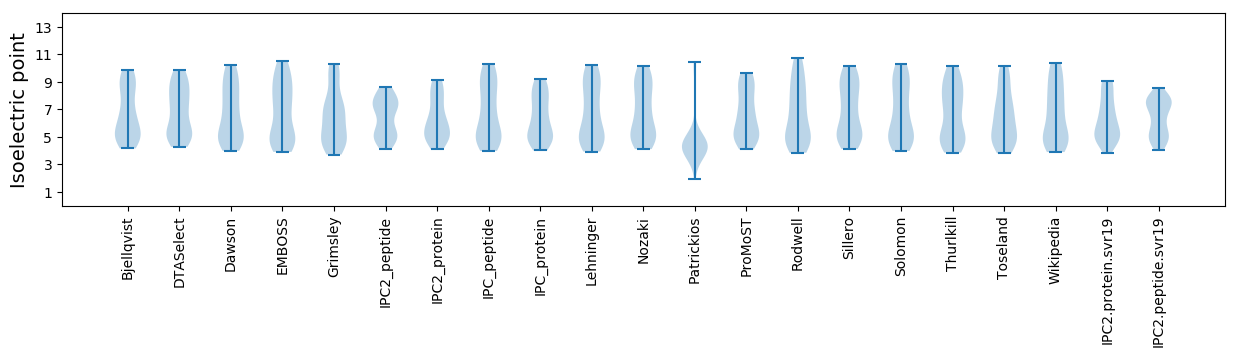
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A059VA49|A0A059VA49_9CAUD DNA restriction methylase OS=Pseudomonas phage phiPSA1 OX=1500757 PE=4 SV=1
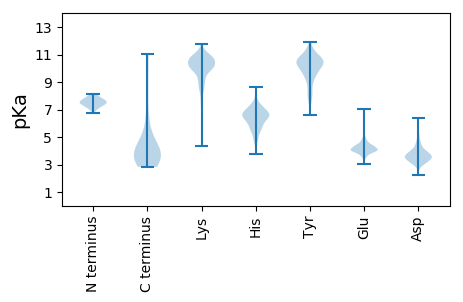
MM1 pKa = 7.83PEE3 pKa = 3.94EE4 pKa = 4.14KK5 pKa = 10.57LIGPVEE11 pKa = 4.11VTRR14 pKa = 11.84DD15 pKa = 3.44EE16 pKa = 4.09NGYY19 pKa = 8.86WYY21 pKa = 10.55HH22 pKa = 6.81PGIPNFDD29 pKa = 4.0EE30 pKa = 4.42DD31 pKa = 3.67HH32 pKa = 6.61AAYY35 pKa = 9.46KK36 pKa = 10.49AWLDD40 pKa = 3.77GQQLEE45 pKa = 4.68VVGWRR50 pKa = 11.84MEE52 pKa = 4.02ADD54 pKa = 4.06LEE56 pKa = 4.36SHH58 pKa = 7.2PYY60 pKa = 8.55WEE62 pKa = 4.71EE63 pKa = 3.62GAANCLGWEE72 pKa = 4.17PEE74 pKa = 4.4KK75 pKa = 10.94PPAYY79 pKa = 10.24DD80 pKa = 3.05WFLLGIFDD88 pKa = 4.66TDD90 pKa = 3.82DD91 pKa = 3.53GPYY94 pKa = 9.29VQWAQRR100 pKa = 11.84VVTPP104 pKa = 4.74
MM1 pKa = 7.83PEE3 pKa = 3.94EE4 pKa = 4.14KK5 pKa = 10.57LIGPVEE11 pKa = 4.11VTRR14 pKa = 11.84DD15 pKa = 3.44EE16 pKa = 4.09NGYY19 pKa = 8.86WYY21 pKa = 10.55HH22 pKa = 6.81PGIPNFDD29 pKa = 4.0EE30 pKa = 4.42DD31 pKa = 3.67HH32 pKa = 6.61AAYY35 pKa = 9.46KK36 pKa = 10.49AWLDD40 pKa = 3.77GQQLEE45 pKa = 4.68VVGWRR50 pKa = 11.84MEE52 pKa = 4.02ADD54 pKa = 4.06LEE56 pKa = 4.36SHH58 pKa = 7.2PYY60 pKa = 8.55WEE62 pKa = 4.71EE63 pKa = 3.62GAANCLGWEE72 pKa = 4.17PEE74 pKa = 4.4KK75 pKa = 10.94PPAYY79 pKa = 10.24DD80 pKa = 3.05WFLLGIFDD88 pKa = 4.66TDD90 pKa = 3.82DD91 pKa = 3.53GPYY94 pKa = 9.29VQWAQRR100 pKa = 11.84VVTPP104 pKa = 4.74
Molecular weight: 12.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A059VJV7|A0A059VJV7_9CAUD Uncharacterized protein OS=Pseudomonas phage phiPSA1 OX=1500757 PE=4 SV=1
MM1 pKa = 7.37KK2 pKa = 10.15AAEE5 pKa = 4.11VLQAIEE11 pKa = 4.21GLEE14 pKa = 3.96PAAQRR19 pKa = 11.84AYY21 pKa = 10.42LAQIAQSLDD30 pKa = 3.58SVSLAEE36 pKa = 4.08VEE38 pKa = 4.19RR39 pKa = 11.84AIDD42 pKa = 3.68SGDD45 pKa = 3.3EE46 pKa = 4.27SAVVAAVQLGIFAGLVEE63 pKa = 4.81HH64 pKa = 7.24LRR66 pKa = 11.84TAYY69 pKa = 10.64VKK71 pKa = 10.57GAQTEE76 pKa = 4.02AAGIKK81 pKa = 10.17VKK83 pKa = 10.86GISKK87 pKa = 10.13EE88 pKa = 4.07LDD90 pKa = 3.21FHH92 pKa = 7.72ASGSAGFMAEE102 pKa = 3.83QASRR106 pKa = 11.84LVAQASADD114 pKa = 3.43QVVAVRR120 pKa = 11.84AVMAYY125 pKa = 10.08GSASGQSARR134 pKa = 11.84KK135 pKa = 8.03MALDD139 pKa = 4.37LIGRR143 pKa = 11.84ISKK146 pKa = 8.06QTGQRR151 pKa = 11.84TGGVLGLSGGYY162 pKa = 9.86AEE164 pKa = 5.33SVILAKK170 pKa = 9.52TQLLSGEE177 pKa = 4.04KK178 pKa = 10.6AMLRR182 pKa = 11.84RR183 pKa = 11.84YY184 pKa = 9.49LLRR187 pKa = 11.84VRR189 pKa = 11.84RR190 pKa = 11.84DD191 pKa = 2.96RR192 pKa = 11.84RR193 pKa = 11.84FDD195 pKa = 3.29PTVRR199 pKa = 11.84AAIKK203 pKa = 10.19SGKK206 pKa = 8.97PLDD209 pKa = 3.85EE210 pKa = 4.1EE211 pKa = 4.52AVNKK215 pKa = 10.03IAGRR219 pKa = 11.84YY220 pKa = 8.74ADD222 pKa = 4.96RR223 pKa = 11.84LLATQAEE230 pKa = 4.78MVAQTFVAEE239 pKa = 4.37SFNEE243 pKa = 3.86GRR245 pKa = 11.84DD246 pKa = 3.62QAWRR250 pKa = 11.84QVVARR255 pKa = 11.84SKK257 pKa = 11.1GRR259 pKa = 11.84LSFIKK264 pKa = 8.6TWKK267 pKa = 10.02SRR269 pKa = 11.84GDD271 pKa = 3.34GKK273 pKa = 10.92VRR275 pKa = 11.84YY276 pKa = 9.15SHH278 pKa = 7.12IAMNGQQVDD287 pKa = 3.24KK288 pKa = 10.07DD289 pKa = 3.79QPFVSPRR296 pKa = 11.84GALLMFPCDD305 pKa = 3.74SSLGAPLSEE314 pKa = 4.53RR315 pKa = 11.84ARR317 pKa = 11.84CRR319 pKa = 11.84CTAEE323 pKa = 3.7YY324 pKa = 10.69SIVQLRR330 pKa = 11.84II331 pKa = 3.16
MM1 pKa = 7.37KK2 pKa = 10.15AAEE5 pKa = 4.11VLQAIEE11 pKa = 4.21GLEE14 pKa = 3.96PAAQRR19 pKa = 11.84AYY21 pKa = 10.42LAQIAQSLDD30 pKa = 3.58SVSLAEE36 pKa = 4.08VEE38 pKa = 4.19RR39 pKa = 11.84AIDD42 pKa = 3.68SGDD45 pKa = 3.3EE46 pKa = 4.27SAVVAAVQLGIFAGLVEE63 pKa = 4.81HH64 pKa = 7.24LRR66 pKa = 11.84TAYY69 pKa = 10.64VKK71 pKa = 10.57GAQTEE76 pKa = 4.02AAGIKK81 pKa = 10.17VKK83 pKa = 10.86GISKK87 pKa = 10.13EE88 pKa = 4.07LDD90 pKa = 3.21FHH92 pKa = 7.72ASGSAGFMAEE102 pKa = 3.83QASRR106 pKa = 11.84LVAQASADD114 pKa = 3.43QVVAVRR120 pKa = 11.84AVMAYY125 pKa = 10.08GSASGQSARR134 pKa = 11.84KK135 pKa = 8.03MALDD139 pKa = 4.37LIGRR143 pKa = 11.84ISKK146 pKa = 8.06QTGQRR151 pKa = 11.84TGGVLGLSGGYY162 pKa = 9.86AEE164 pKa = 5.33SVILAKK170 pKa = 9.52TQLLSGEE177 pKa = 4.04KK178 pKa = 10.6AMLRR182 pKa = 11.84RR183 pKa = 11.84YY184 pKa = 9.49LLRR187 pKa = 11.84VRR189 pKa = 11.84RR190 pKa = 11.84DD191 pKa = 2.96RR192 pKa = 11.84RR193 pKa = 11.84FDD195 pKa = 3.29PTVRR199 pKa = 11.84AAIKK203 pKa = 10.19SGKK206 pKa = 8.97PLDD209 pKa = 3.85EE210 pKa = 4.1EE211 pKa = 4.52AVNKK215 pKa = 10.03IAGRR219 pKa = 11.84YY220 pKa = 8.74ADD222 pKa = 4.96RR223 pKa = 11.84LLATQAEE230 pKa = 4.78MVAQTFVAEE239 pKa = 4.37SFNEE243 pKa = 3.86GRR245 pKa = 11.84DD246 pKa = 3.62QAWRR250 pKa = 11.84QVVARR255 pKa = 11.84SKK257 pKa = 11.1GRR259 pKa = 11.84LSFIKK264 pKa = 8.6TWKK267 pKa = 10.02SRR269 pKa = 11.84GDD271 pKa = 3.34GKK273 pKa = 10.92VRR275 pKa = 11.84YY276 pKa = 9.15SHH278 pKa = 7.12IAMNGQQVDD287 pKa = 3.24KK288 pKa = 10.07DD289 pKa = 3.79QPFVSPRR296 pKa = 11.84GALLMFPCDD305 pKa = 3.74SSLGAPLSEE314 pKa = 4.53RR315 pKa = 11.84ARR317 pKa = 11.84CRR319 pKa = 11.84CTAEE323 pKa = 3.7YY324 pKa = 10.69SIVQLRR330 pKa = 11.84II331 pKa = 3.16
Molecular weight: 35.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13753 |
70 |
1344 |
264.5 |
28.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.121 ± 0.711 | 1.083 ± 0.149 |
5.737 ± 0.272 | 5.941 ± 0.274 |
3.294 ± 0.206 | 7.991 ± 0.348 |
1.614 ± 0.235 | 5.003 ± 0.198 |
4.893 ± 0.359 | 7.657 ± 0.314 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.341 ± 0.146 | 3.752 ± 0.271 |
4.094 ± 0.31 | 4.588 ± 0.375 |
5.89 ± 0.346 | 6.689 ± 0.356 |
6.071 ± 0.268 | 6.784 ± 0.244 |
1.578 ± 0.198 | 2.879 ± 0.15 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
