
Tortoise microvirus 47
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
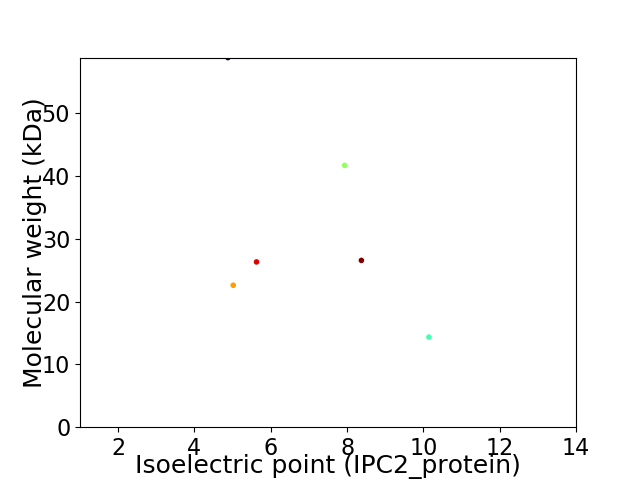
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVY9|A0A4V1FVY9_9VIRU Uncharacterized protein OS=Tortoise microvirus 47 OX=2583151 PE=4 SV=1
MM1 pKa = 7.7RR2 pKa = 11.84RR3 pKa = 11.84QSARR7 pKa = 11.84GITSLPAGKK16 pKa = 8.49MVPIAAFPLLRR27 pKa = 11.84EE28 pKa = 3.94DD29 pKa = 4.99AIRR32 pKa = 11.84SGRR35 pKa = 11.84LRR37 pKa = 11.84FSFEE41 pKa = 3.98SMEE44 pKa = 4.11TAEE47 pKa = 4.51ILMNAINVCVMAYY60 pKa = 9.8LVPNLALEE68 pKa = 4.31RR69 pKa = 11.84FNGSMDD75 pKa = 3.71QLNLSYY81 pKa = 10.44EE82 pKa = 4.33GKK84 pKa = 9.65PPLDD88 pKa = 4.11GEE90 pKa = 4.45PVTPYY95 pKa = 10.43IEE97 pKa = 4.08TAPFGAHH104 pKa = 5.56GTNDD108 pKa = 2.59IYY110 pKa = 11.08TYY112 pKa = 10.7LGLHH116 pKa = 6.34GRR118 pKa = 11.84PTQQVNTAYY127 pKa = 10.85LEE129 pKa = 4.4AYY131 pKa = 8.74NCIWNFRR138 pKa = 11.84AKK140 pKa = 10.35NRR142 pKa = 11.84SKK144 pKa = 10.61EE145 pKa = 3.8LSEE148 pKa = 4.05RR149 pKa = 11.84ARR151 pKa = 11.84LQTDD155 pKa = 4.22LAPAFWSHH163 pKa = 7.31DD164 pKa = 3.76MFKK167 pKa = 10.67HH168 pKa = 5.57IVPDD172 pKa = 4.2FDD174 pKa = 3.28QAAIDD179 pKa = 4.41GEE181 pKa = 4.69VPLNITDD188 pKa = 2.68GRR190 pKa = 11.84MPVRR194 pKa = 11.84GIGLGGTTSIATNSAVRR211 pKa = 11.84EE212 pKa = 4.25SGGTATYY219 pKa = 10.89ARR221 pKa = 11.84STTTAVDD228 pKa = 3.46TGQSVRR234 pKa = 11.84LQLAATGTGTTVLPDD249 pKa = 3.34IYY251 pKa = 11.34AEE253 pKa = 4.04MEE255 pKa = 4.15EE256 pKa = 4.11NGITVSLSNIEE267 pKa = 4.22LARR270 pKa = 11.84KK271 pKa = 6.35TQAFAKK277 pKa = 10.47LRR279 pKa = 11.84TQYY282 pKa = 10.49NAHH285 pKa = 7.39DD286 pKa = 3.67DD287 pKa = 4.12WIINLLMDD295 pKa = 5.67GISIPEE301 pKa = 3.87QAFKK305 pKa = 11.11QPMLLAEE312 pKa = 4.29KK313 pKa = 9.66RR314 pKa = 11.84TIFGMAKK321 pKa = 9.85RR322 pKa = 11.84YY323 pKa = 10.23ASDD326 pKa = 3.33SGNLTEE332 pKa = 5.04SVVNGATFLDD342 pKa = 4.23MSIQLPRR349 pKa = 11.84IGTGGVVMIVAEE361 pKa = 4.15ITPDD365 pKa = 3.3QLFEE369 pKa = 4.15RR370 pKa = 11.84QEE372 pKa = 3.86DD373 pKa = 4.71PYY375 pKa = 11.75LHH377 pKa = 5.67TTNVEE382 pKa = 3.78QLPQYY387 pKa = 10.93LRR389 pKa = 11.84DD390 pKa = 3.58TLDD393 pKa = 3.47PEE395 pKa = 4.42KK396 pKa = 11.1VDD398 pKa = 3.03IVKK401 pKa = 10.6NGRR404 pKa = 11.84IDD406 pKa = 3.54VDD408 pKa = 3.37HH409 pKa = 7.37DD410 pKa = 3.75QPDD413 pKa = 3.28ATFGYY418 pKa = 10.29EE419 pKa = 3.87PLNARR424 pKa = 11.84WNINAPRR431 pKa = 11.84IGARR435 pKa = 11.84FYY437 pKa = 10.94RR438 pKa = 11.84PEE440 pKa = 3.54VDD442 pKa = 4.19AGFDD446 pKa = 3.54EE447 pKa = 4.48DD448 pKa = 4.37RR449 pKa = 11.84QRR451 pKa = 11.84IWAVEE456 pKa = 4.24TKK458 pKa = 10.68NPTLSTDD465 pKa = 5.09FYY467 pKa = 11.08LCTNMHH473 pKa = 6.47TKK475 pKa = 10.32PFVVTNQDD483 pKa = 2.92PFEE486 pKa = 4.31VVTQGDD492 pKa = 3.9VFIEE496 pKa = 4.52GNTVFGGHH504 pKa = 7.35LIEE507 pKa = 5.26ATDD510 pKa = 4.15DD511 pKa = 3.9YY512 pKa = 11.6EE513 pKa = 4.91KK514 pKa = 11.21VLAVADD520 pKa = 3.48QTRR523 pKa = 11.84IEE525 pKa = 4.28KK526 pKa = 9.86EE527 pKa = 3.86QAA529 pKa = 2.85
MM1 pKa = 7.7RR2 pKa = 11.84RR3 pKa = 11.84QSARR7 pKa = 11.84GITSLPAGKK16 pKa = 8.49MVPIAAFPLLRR27 pKa = 11.84EE28 pKa = 3.94DD29 pKa = 4.99AIRR32 pKa = 11.84SGRR35 pKa = 11.84LRR37 pKa = 11.84FSFEE41 pKa = 3.98SMEE44 pKa = 4.11TAEE47 pKa = 4.51ILMNAINVCVMAYY60 pKa = 9.8LVPNLALEE68 pKa = 4.31RR69 pKa = 11.84FNGSMDD75 pKa = 3.71QLNLSYY81 pKa = 10.44EE82 pKa = 4.33GKK84 pKa = 9.65PPLDD88 pKa = 4.11GEE90 pKa = 4.45PVTPYY95 pKa = 10.43IEE97 pKa = 4.08TAPFGAHH104 pKa = 5.56GTNDD108 pKa = 2.59IYY110 pKa = 11.08TYY112 pKa = 10.7LGLHH116 pKa = 6.34GRR118 pKa = 11.84PTQQVNTAYY127 pKa = 10.85LEE129 pKa = 4.4AYY131 pKa = 8.74NCIWNFRR138 pKa = 11.84AKK140 pKa = 10.35NRR142 pKa = 11.84SKK144 pKa = 10.61EE145 pKa = 3.8LSEE148 pKa = 4.05RR149 pKa = 11.84ARR151 pKa = 11.84LQTDD155 pKa = 4.22LAPAFWSHH163 pKa = 7.31DD164 pKa = 3.76MFKK167 pKa = 10.67HH168 pKa = 5.57IVPDD172 pKa = 4.2FDD174 pKa = 3.28QAAIDD179 pKa = 4.41GEE181 pKa = 4.69VPLNITDD188 pKa = 2.68GRR190 pKa = 11.84MPVRR194 pKa = 11.84GIGLGGTTSIATNSAVRR211 pKa = 11.84EE212 pKa = 4.25SGGTATYY219 pKa = 10.89ARR221 pKa = 11.84STTTAVDD228 pKa = 3.46TGQSVRR234 pKa = 11.84LQLAATGTGTTVLPDD249 pKa = 3.34IYY251 pKa = 11.34AEE253 pKa = 4.04MEE255 pKa = 4.15EE256 pKa = 4.11NGITVSLSNIEE267 pKa = 4.22LARR270 pKa = 11.84KK271 pKa = 6.35TQAFAKK277 pKa = 10.47LRR279 pKa = 11.84TQYY282 pKa = 10.49NAHH285 pKa = 7.39DD286 pKa = 3.67DD287 pKa = 4.12WIINLLMDD295 pKa = 5.67GISIPEE301 pKa = 3.87QAFKK305 pKa = 11.11QPMLLAEE312 pKa = 4.29KK313 pKa = 9.66RR314 pKa = 11.84TIFGMAKK321 pKa = 9.85RR322 pKa = 11.84YY323 pKa = 10.23ASDD326 pKa = 3.33SGNLTEE332 pKa = 5.04SVVNGATFLDD342 pKa = 4.23MSIQLPRR349 pKa = 11.84IGTGGVVMIVAEE361 pKa = 4.15ITPDD365 pKa = 3.3QLFEE369 pKa = 4.15RR370 pKa = 11.84QEE372 pKa = 3.86DD373 pKa = 4.71PYY375 pKa = 11.75LHH377 pKa = 5.67TTNVEE382 pKa = 3.78QLPQYY387 pKa = 10.93LRR389 pKa = 11.84DD390 pKa = 3.58TLDD393 pKa = 3.47PEE395 pKa = 4.42KK396 pKa = 11.1VDD398 pKa = 3.03IVKK401 pKa = 10.6NGRR404 pKa = 11.84IDD406 pKa = 3.54VDD408 pKa = 3.37HH409 pKa = 7.37DD410 pKa = 3.75QPDD413 pKa = 3.28ATFGYY418 pKa = 10.29EE419 pKa = 3.87PLNARR424 pKa = 11.84WNINAPRR431 pKa = 11.84IGARR435 pKa = 11.84FYY437 pKa = 10.94RR438 pKa = 11.84PEE440 pKa = 3.54VDD442 pKa = 4.19AGFDD446 pKa = 3.54EE447 pKa = 4.48DD448 pKa = 4.37RR449 pKa = 11.84QRR451 pKa = 11.84IWAVEE456 pKa = 4.24TKK458 pKa = 10.68NPTLSTDD465 pKa = 5.09FYY467 pKa = 11.08LCTNMHH473 pKa = 6.47TKK475 pKa = 10.32PFVVTNQDD483 pKa = 2.92PFEE486 pKa = 4.31VVTQGDD492 pKa = 3.9VFIEE496 pKa = 4.52GNTVFGGHH504 pKa = 7.35LIEE507 pKa = 5.26ATDD510 pKa = 4.15DD511 pKa = 3.9YY512 pKa = 11.6EE513 pKa = 4.91KK514 pKa = 11.21VLAVADD520 pKa = 3.48QTRR523 pKa = 11.84IEE525 pKa = 4.28KK526 pKa = 9.86EE527 pKa = 3.86QAA529 pKa = 2.85
Molecular weight: 58.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVY9|A0A4V1FVY9_9VIRU Uncharacterized protein OS=Tortoise microvirus 47 OX=2583151 PE=4 SV=1
MM1 pKa = 7.27ARR3 pKa = 11.84KK4 pKa = 9.47NRR6 pKa = 11.84TRR8 pKa = 11.84RR9 pKa = 11.84SKK11 pKa = 10.64AAASRR16 pKa = 11.84GAKK19 pKa = 9.15GKK21 pKa = 9.76NAASVQNRR29 pKa = 11.84DD30 pKa = 3.28ARR32 pKa = 11.84SLAASPLAFMRR43 pKa = 11.84AATIKK48 pKa = 10.63SPTRR52 pKa = 11.84AKK54 pKa = 10.3SVNVPRR60 pKa = 11.84RR61 pKa = 11.84QIVEE65 pKa = 3.96IEE67 pKa = 4.64LPPAPIIQEE76 pKa = 3.98NKK78 pKa = 8.9RR79 pKa = 11.84ARR81 pKa = 11.84SALALKK87 pKa = 10.38AISEE91 pKa = 4.06PDD93 pKa = 3.32NRR95 pKa = 11.84KK96 pKa = 9.7SSPKK100 pKa = 10.15ARR102 pKa = 11.84EE103 pKa = 3.53QLHH106 pKa = 5.91CKK108 pKa = 9.61KK109 pKa = 10.7RR110 pKa = 11.84PDD112 pKa = 3.53SKK114 pKa = 10.92KK115 pKa = 10.13AATGKK120 pKa = 10.58GGGVKK125 pKa = 10.22RR126 pKa = 11.84FIPWCGG132 pKa = 2.95
MM1 pKa = 7.27ARR3 pKa = 11.84KK4 pKa = 9.47NRR6 pKa = 11.84TRR8 pKa = 11.84RR9 pKa = 11.84SKK11 pKa = 10.64AAASRR16 pKa = 11.84GAKK19 pKa = 9.15GKK21 pKa = 9.76NAASVQNRR29 pKa = 11.84DD30 pKa = 3.28ARR32 pKa = 11.84SLAASPLAFMRR43 pKa = 11.84AATIKK48 pKa = 10.63SPTRR52 pKa = 11.84AKK54 pKa = 10.3SVNVPRR60 pKa = 11.84RR61 pKa = 11.84QIVEE65 pKa = 3.96IEE67 pKa = 4.64LPPAPIIQEE76 pKa = 3.98NKK78 pKa = 8.9RR79 pKa = 11.84ARR81 pKa = 11.84SALALKK87 pKa = 10.38AISEE91 pKa = 4.06PDD93 pKa = 3.32NRR95 pKa = 11.84KK96 pKa = 9.7SSPKK100 pKa = 10.15ARR102 pKa = 11.84EE103 pKa = 3.53QLHH106 pKa = 5.91CKK108 pKa = 9.61KK109 pKa = 10.7RR110 pKa = 11.84PDD112 pKa = 3.53SKK114 pKa = 10.92KK115 pKa = 10.13AATGKK120 pKa = 10.58GGGVKK125 pKa = 10.22RR126 pKa = 11.84FIPWCGG132 pKa = 2.95
Molecular weight: 14.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1702 |
132 |
529 |
283.7 |
31.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.694 ± 1.091 | 0.881 ± 0.301 |
5.464 ± 0.681 | 6.816 ± 0.568 |
3.584 ± 0.34 | 8.049 ± 0.677 |
2.115 ± 0.376 | 4.524 ± 0.614 |
4.994 ± 1.112 | 6.404 ± 0.645 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.703 ± 0.166 | 4.348 ± 0.461 |
6.345 ± 0.354 | 4.583 ± 0.262 |
7.462 ± 0.886 | 5.112 ± 0.819 |
5.699 ± 1.08 | 5.993 ± 0.69 |
1.939 ± 0.542 | 3.29 ± 0.858 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
