
Chicken stool-associated circular virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
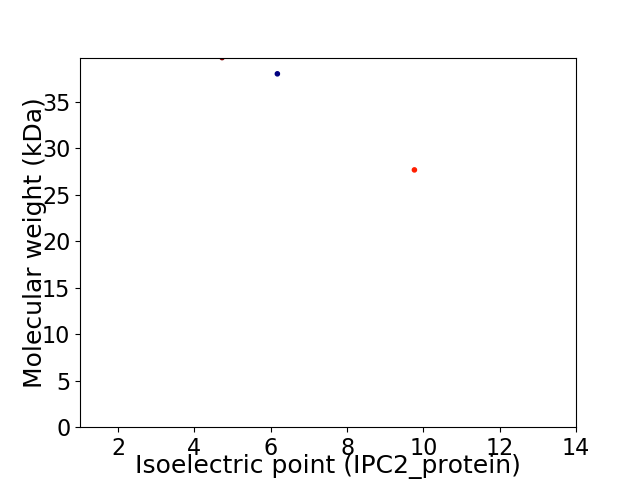
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
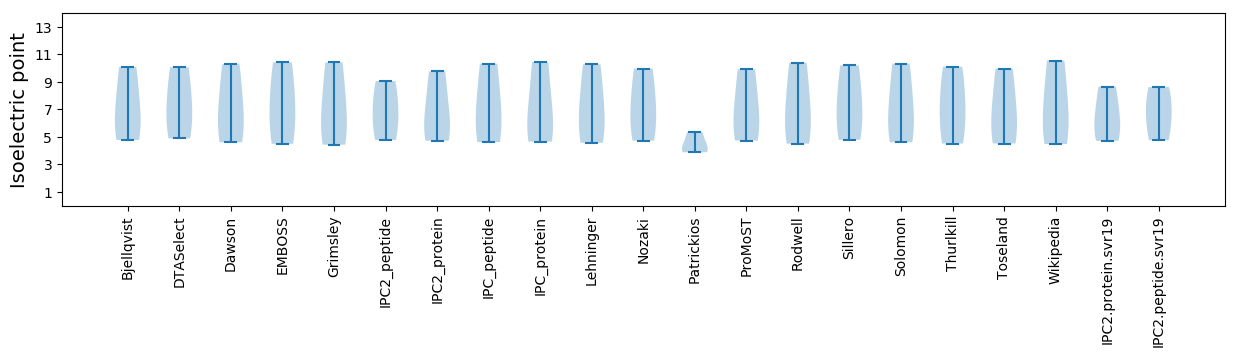
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L6KW42|A0A1L6KW42_9VIRU Replication associated protein OS=Chicken stool-associated circular virus OX=1930302 PE=3 SV=1
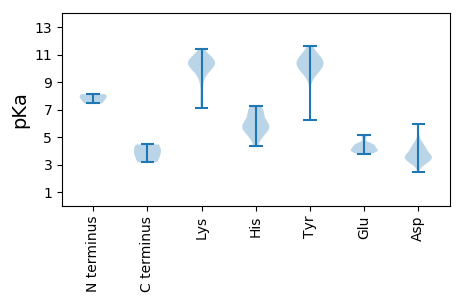
MM1 pKa = 7.44SWNKK5 pKa = 7.13YY6 pKa = 6.24TKK8 pKa = 9.96PLFLTCEE15 pKa = 5.09DD16 pKa = 3.39ITSGPRR22 pKa = 11.84VWIPPRR28 pKa = 11.84GMAYY32 pKa = 10.28SRR34 pKa = 11.84VILMSNGTDD43 pKa = 4.42RR44 pKa = 11.84PYY46 pKa = 10.68PWYY49 pKa = 10.01DD50 pKa = 2.94NKK52 pKa = 10.94LYY54 pKa = 10.29EE55 pKa = 4.52IKK57 pKa = 10.74DD58 pKa = 3.61EE59 pKa = 4.52GEE61 pKa = 4.03HH62 pKa = 5.93VLLEE66 pKa = 4.34RR67 pKa = 11.84VDD69 pKa = 3.56EE70 pKa = 4.54GRR72 pKa = 11.84PIMLYY77 pKa = 9.57RR78 pKa = 11.84PRR80 pKa = 11.84VIIDD84 pKa = 3.38TSKK87 pKa = 10.55HH88 pKa = 4.34YY89 pKa = 9.95QTMEE93 pKa = 3.92KK94 pKa = 9.57TNKK97 pKa = 8.24TVYY100 pKa = 10.42PLVEE104 pKa = 4.45EE105 pKa = 4.41YY106 pKa = 11.15NLLGTVPKK114 pKa = 10.43TNVTITNDD122 pKa = 3.01IKK124 pKa = 11.11DD125 pKa = 4.26EE126 pKa = 4.08KK127 pKa = 10.56WPDD130 pKa = 3.64EE131 pKa = 5.13DD132 pKa = 4.94PDD134 pKa = 4.08NPDD137 pKa = 3.5EE138 pKa = 4.51INRR141 pKa = 11.84DD142 pKa = 3.8FPPYY146 pKa = 10.07GYY148 pKa = 10.8SKK150 pKa = 9.84VTTEE154 pKa = 4.18IKK156 pKa = 10.14WNGEE160 pKa = 3.79DD161 pKa = 5.98FPIHH165 pKa = 5.82WLEE168 pKa = 3.8QKK170 pKa = 10.16EE171 pKa = 4.16YY172 pKa = 10.11NLEE175 pKa = 4.15KK176 pKa = 10.34EE177 pKa = 4.09TDD179 pKa = 3.62EE180 pKa = 4.43NGITIDD186 pKa = 3.33EE187 pKa = 4.39WWRR190 pKa = 11.84EE191 pKa = 4.03TAIEE195 pKa = 4.26GADD198 pKa = 3.4NIYY201 pKa = 10.97YY202 pKa = 10.46CGLKK206 pKa = 9.29MNVVIRR212 pKa = 11.84ISVSAINRR220 pKa = 11.84EE221 pKa = 4.04SLDD224 pKa = 3.42TFKK227 pKa = 11.12FSLAGGEE234 pKa = 3.84ILQPGEE240 pKa = 3.92SGIFIQRR247 pKa = 11.84TLDD250 pKa = 3.4VDD252 pKa = 4.18SSPVEE257 pKa = 3.9YY258 pKa = 10.64QITLVYY264 pKa = 11.15NNTNEE269 pKa = 3.82IKK271 pKa = 9.78TVFKK275 pKa = 10.39PVSQTRR281 pKa = 11.84HH282 pKa = 5.39ALLDD286 pKa = 3.81FSDD289 pKa = 4.47GYY291 pKa = 10.53KK292 pKa = 10.13IKK294 pKa = 10.53IKK296 pKa = 10.58AGHH299 pKa = 6.91DD300 pKa = 2.98KK301 pKa = 11.41DD302 pKa = 3.8MIEE305 pKa = 3.97IQGDD309 pKa = 3.85VTTLNIGSGLPEE321 pKa = 3.97ITCSLSAKK329 pKa = 7.71MFSMIFGNNWKK340 pKa = 9.73PDD342 pKa = 3.62DD343 pKa = 3.88EE344 pKa = 4.46
MM1 pKa = 7.44SWNKK5 pKa = 7.13YY6 pKa = 6.24TKK8 pKa = 9.96PLFLTCEE15 pKa = 5.09DD16 pKa = 3.39ITSGPRR22 pKa = 11.84VWIPPRR28 pKa = 11.84GMAYY32 pKa = 10.28SRR34 pKa = 11.84VILMSNGTDD43 pKa = 4.42RR44 pKa = 11.84PYY46 pKa = 10.68PWYY49 pKa = 10.01DD50 pKa = 2.94NKK52 pKa = 10.94LYY54 pKa = 10.29EE55 pKa = 4.52IKK57 pKa = 10.74DD58 pKa = 3.61EE59 pKa = 4.52GEE61 pKa = 4.03HH62 pKa = 5.93VLLEE66 pKa = 4.34RR67 pKa = 11.84VDD69 pKa = 3.56EE70 pKa = 4.54GRR72 pKa = 11.84PIMLYY77 pKa = 9.57RR78 pKa = 11.84PRR80 pKa = 11.84VIIDD84 pKa = 3.38TSKK87 pKa = 10.55HH88 pKa = 4.34YY89 pKa = 9.95QTMEE93 pKa = 3.92KK94 pKa = 9.57TNKK97 pKa = 8.24TVYY100 pKa = 10.42PLVEE104 pKa = 4.45EE105 pKa = 4.41YY106 pKa = 11.15NLLGTVPKK114 pKa = 10.43TNVTITNDD122 pKa = 3.01IKK124 pKa = 11.11DD125 pKa = 4.26EE126 pKa = 4.08KK127 pKa = 10.56WPDD130 pKa = 3.64EE131 pKa = 5.13DD132 pKa = 4.94PDD134 pKa = 4.08NPDD137 pKa = 3.5EE138 pKa = 4.51INRR141 pKa = 11.84DD142 pKa = 3.8FPPYY146 pKa = 10.07GYY148 pKa = 10.8SKK150 pKa = 9.84VTTEE154 pKa = 4.18IKK156 pKa = 10.14WNGEE160 pKa = 3.79DD161 pKa = 5.98FPIHH165 pKa = 5.82WLEE168 pKa = 3.8QKK170 pKa = 10.16EE171 pKa = 4.16YY172 pKa = 10.11NLEE175 pKa = 4.15KK176 pKa = 10.34EE177 pKa = 4.09TDD179 pKa = 3.62EE180 pKa = 4.43NGITIDD186 pKa = 3.33EE187 pKa = 4.39WWRR190 pKa = 11.84EE191 pKa = 4.03TAIEE195 pKa = 4.26GADD198 pKa = 3.4NIYY201 pKa = 10.97YY202 pKa = 10.46CGLKK206 pKa = 9.29MNVVIRR212 pKa = 11.84ISVSAINRR220 pKa = 11.84EE221 pKa = 4.04SLDD224 pKa = 3.42TFKK227 pKa = 11.12FSLAGGEE234 pKa = 3.84ILQPGEE240 pKa = 3.92SGIFIQRR247 pKa = 11.84TLDD250 pKa = 3.4VDD252 pKa = 4.18SSPVEE257 pKa = 3.9YY258 pKa = 10.64QITLVYY264 pKa = 11.15NNTNEE269 pKa = 3.82IKK271 pKa = 9.78TVFKK275 pKa = 10.39PVSQTRR281 pKa = 11.84HH282 pKa = 5.39ALLDD286 pKa = 3.81FSDD289 pKa = 4.47GYY291 pKa = 10.53KK292 pKa = 10.13IKK294 pKa = 10.53IKK296 pKa = 10.58AGHH299 pKa = 6.91DD300 pKa = 2.98KK301 pKa = 11.41DD302 pKa = 3.8MIEE305 pKa = 3.97IQGDD309 pKa = 3.85VTTLNIGSGLPEE321 pKa = 3.97ITCSLSAKK329 pKa = 7.71MFSMIFGNNWKK340 pKa = 9.73PDD342 pKa = 3.62DD343 pKa = 3.88EE344 pKa = 4.46
Molecular weight: 39.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L6KW76|A0A1L6KW76_9VIRU Capsid protein OS=Chicken stool-associated circular virus OX=1930302 PE=4 SV=1
MM1 pKa = 7.9AYY3 pKa = 10.02ASSYY7 pKa = 10.44FRR9 pKa = 11.84TRR11 pKa = 11.84YY12 pKa = 9.45ASYY15 pKa = 10.45RR16 pKa = 11.84RR17 pKa = 11.84AARR20 pKa = 11.84GTRR23 pKa = 11.84RR24 pKa = 11.84WNSGSRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84AWGNARR38 pKa = 11.84AARR41 pKa = 11.84QQRR44 pKa = 11.84DD45 pKa = 3.25ACTVTISRR53 pKa = 11.84LTNVSVPVAASQTSGTAFIAHH74 pKa = 7.24WDD76 pKa = 3.63SLRR79 pKa = 11.84QSTYY83 pKa = 9.36FPNYY87 pKa = 9.79SSMYY91 pKa = 10.05DD92 pKa = 3.12QIKK95 pKa = 9.5IDD97 pKa = 3.88KK98 pKa = 10.19IRR100 pKa = 11.84VKK102 pKa = 10.11VTGTNAGASTDD113 pKa = 3.9LVSPTVVLAFDD124 pKa = 4.53RR125 pKa = 11.84NGLDD129 pKa = 3.41SAQLNSDD136 pKa = 4.71GISLATGVQGVSTYY150 pKa = 11.22SSAQSRR156 pKa = 11.84QWSVGNAFTMWQTIYY171 pKa = 10.49PSTIMEE177 pKa = 4.44KK178 pKa = 10.52GQYY181 pKa = 9.73IPTNSLSTSSTTANPAYY198 pKa = 10.19VISDD202 pKa = 3.12STMPFKK208 pKa = 10.63PVTLFGISTNGTEE221 pKa = 4.19VTSAQTFVFSVEE233 pKa = 3.88LEE235 pKa = 4.12YY236 pKa = 10.73TVTFRR241 pKa = 11.84GMRR244 pKa = 11.84KK245 pKa = 9.53PEE247 pKa = 3.83IQTGSS252 pKa = 3.16
MM1 pKa = 7.9AYY3 pKa = 10.02ASSYY7 pKa = 10.44FRR9 pKa = 11.84TRR11 pKa = 11.84YY12 pKa = 9.45ASYY15 pKa = 10.45RR16 pKa = 11.84RR17 pKa = 11.84AARR20 pKa = 11.84GTRR23 pKa = 11.84RR24 pKa = 11.84WNSGSRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84AWGNARR38 pKa = 11.84AARR41 pKa = 11.84QQRR44 pKa = 11.84DD45 pKa = 3.25ACTVTISRR53 pKa = 11.84LTNVSVPVAASQTSGTAFIAHH74 pKa = 7.24WDD76 pKa = 3.63SLRR79 pKa = 11.84QSTYY83 pKa = 9.36FPNYY87 pKa = 9.79SSMYY91 pKa = 10.05DD92 pKa = 3.12QIKK95 pKa = 9.5IDD97 pKa = 3.88KK98 pKa = 10.19IRR100 pKa = 11.84VKK102 pKa = 10.11VTGTNAGASTDD113 pKa = 3.9LVSPTVVLAFDD124 pKa = 4.53RR125 pKa = 11.84NGLDD129 pKa = 3.41SAQLNSDD136 pKa = 4.71GISLATGVQGVSTYY150 pKa = 11.22SSAQSRR156 pKa = 11.84QWSVGNAFTMWQTIYY171 pKa = 10.49PSTIMEE177 pKa = 4.44KK178 pKa = 10.52GQYY181 pKa = 9.73IPTNSLSTSSTTANPAYY198 pKa = 10.19VISDD202 pKa = 3.12STMPFKK208 pKa = 10.63PVTLFGISTNGTEE221 pKa = 4.19VTSAQTFVFSVEE233 pKa = 3.88LEE235 pKa = 4.12YY236 pKa = 10.73TVTFRR241 pKa = 11.84GMRR244 pKa = 11.84KK245 pKa = 9.53PEE247 pKa = 3.83IQTGSS252 pKa = 3.16
Molecular weight: 27.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
923 |
252 |
344 |
307.7 |
35.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.309 ± 1.628 | 1.733 ± 0.899 |
6.717 ± 1.057 | 6.392 ± 1.531 |
3.467 ± 0.271 | 6.717 ± 0.447 |
1.842 ± 0.72 | 6.501 ± 1.214 |
6.826 ± 1.725 | 6.067 ± 0.711 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.925 ± 0.345 | 4.875 ± 0.618 |
4.117 ± 1.036 | 3.359 ± 0.71 |
5.309 ± 0.895 | 6.717 ± 2.401 |
7.909 ± 1.493 | 6.067 ± 0.37 |
2.709 ± 0.32 | 4.442 ± 0.37 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
