
Chrysochromulina tobinii
Taxonomy: cellular organisms; Eukaryota; Haptista; Haptophyta; Prymnesiophyceae; Prymnesiales; Chrysochromulinaceae; Chrysochromulina
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
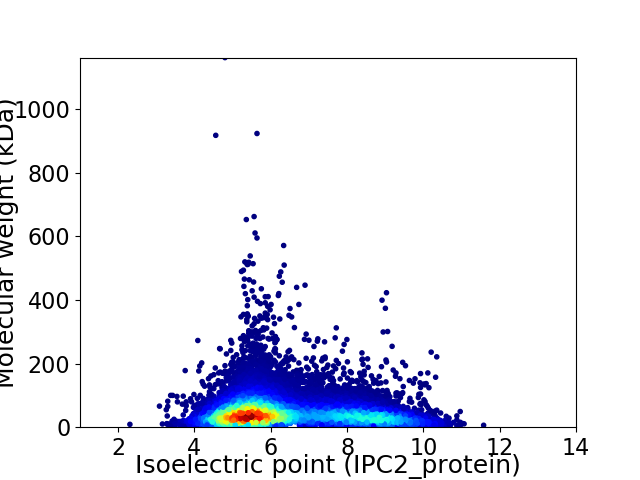
Virtual 2D-PAGE plot for 16679 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M0K1I6|A0A0M0K1I6_9EUKA Coiled coil flagellar protein OS=Chrysochromulina tobinii OX=1460289 GN=Ctob_009394 PE=4 SV=1
MM1 pKa = 7.47SFADD5 pKa = 3.78AQQCATTCIGAPGYY19 pKa = 11.08ASDD22 pKa = 4.75CDD24 pKa = 3.94CAAEE28 pKa = 4.01FASNKK33 pKa = 9.44FATEE37 pKa = 3.7YY38 pKa = 10.52GYY40 pKa = 11.19DD41 pKa = 4.29FITVNGVSSSLTNGPMNVQVATEE64 pKa = 4.15TAQLVDD70 pKa = 3.8TFSTTTFGAVQACTRR85 pKa = 11.84GSLEE89 pKa = 3.96TFKK92 pKa = 11.29ARR94 pKa = 11.84VAAKK98 pKa = 9.84SLSNMTSTGAKK109 pKa = 8.93MDD111 pKa = 3.53PTLNDD116 pKa = 3.05VQTTLSIISYY126 pKa = 10.31LGGLLEE132 pKa = 4.29TDD134 pKa = 3.05AFEE137 pKa = 4.81ANSTIDD143 pKa = 3.12IAEE146 pKa = 4.56AAHH149 pKa = 5.93EE150 pKa = 4.26TDD152 pKa = 4.48PDD154 pKa = 3.99SMPYY158 pKa = 10.36ARR160 pKa = 11.84SSSTGASLVAIAPALLPGMLAVLGLLCAVLYY191 pKa = 10.47GAVQAFAAHH200 pKa = 6.94LGASALASIMGPPPPKK216 pKa = 10.08GRR218 pKa = 11.84RR219 pKa = 11.84AVSAAPASCRR229 pKa = 11.84PSARR233 pKa = 11.84RR234 pKa = 11.84GSAGRR239 pKa = 11.84FGSAPRR245 pKa = 11.84LVGFLLLALFVSATDD260 pKa = 3.77AQCMEE265 pKa = 4.58EE266 pKa = 4.01WCNVCTGEE274 pKa = 4.4CTSATGGEE282 pKa = 4.18LLGSSEE288 pKa = 4.94AVLQFLSSVLPSSSSGLTSSTTFSIATADD317 pKa = 3.39VDD319 pKa = 4.08GDD321 pKa = 3.75GDD323 pKa = 4.97LDD325 pKa = 3.87VLLGGSPTSRR335 pKa = 11.84VLLNAGNGTFPTSIEE350 pKa = 4.1LPGVSAEE357 pKa = 4.19TRR359 pKa = 11.84SIAAADD365 pKa = 3.88LDD367 pKa = 4.21GDD369 pKa = 3.95GDD371 pKa = 4.83LDD373 pKa = 3.87VLLGSSPSSVLLNAGDD389 pKa = 4.03GTFPTSITLPGDD401 pKa = 3.58GALTRR406 pKa = 11.84SITAADD412 pKa = 4.01LDD414 pKa = 4.23GDD416 pKa = 3.94GDD418 pKa = 4.83LDD420 pKa = 3.93VLIGNSGSPSRR431 pKa = 11.84VLLNAGDD438 pKa = 4.08GTFPASIEE446 pKa = 4.12LPGGSPLTYY455 pKa = 10.15SIAAADD461 pKa = 3.63VDD463 pKa = 4.1GDD465 pKa = 3.74GDD467 pKa = 4.8LDD469 pKa = 3.92VLIGNSNSPNRR480 pKa = 11.84VLLNAGDD487 pKa = 4.09GTFPTSITLPGGSAEE502 pKa = 4.36TNSIAAADD510 pKa = 3.73VDD512 pKa = 4.1GDD514 pKa = 3.74GDD516 pKa = 4.8LDD518 pKa = 3.92VLIGNSNSPNRR529 pKa = 11.84VLLNTGNGTFPTSIEE544 pKa = 4.17LPGSIAATQSIAAADD559 pKa = 3.63VDD561 pKa = 4.1GDD563 pKa = 3.77GDD565 pKa = 5.12LDD567 pKa = 3.87VLLGNNGSPSRR578 pKa = 11.84VLLNAGNGTFPTSITLPGGSLFTQSIAAADD608 pKa = 3.63VDD610 pKa = 4.1GDD612 pKa = 3.74GDD614 pKa = 4.8LDD616 pKa = 3.87VLIGNSQYY624 pKa = 10.96RR625 pKa = 11.84NRR627 pKa = 11.84MLLNAGGTFQTIIQLPGPGIVSTHH651 pKa = 6.58SIAAADD657 pKa = 4.29LDD659 pKa = 4.12SDD661 pKa = 3.96GDD663 pKa = 4.15LDD665 pKa = 4.27VLIGNSGSPSQVLLNAGDD683 pKa = 3.99GTFPASITLPGGSPFTQSVAAADD706 pKa = 3.67VDD708 pKa = 3.96GDD710 pKa = 3.74GDD712 pKa = 4.8LDD714 pKa = 3.85VLIGNSYY721 pKa = 10.75SPSLVLLNAGDD732 pKa = 4.08GTFPTSIEE740 pKa = 4.19LPGGTGDD747 pKa = 3.5TRR749 pKa = 11.84SIAAADD755 pKa = 3.54VDD757 pKa = 4.1GDD759 pKa = 3.77GDD761 pKa = 4.83LDD763 pKa = 3.79VLLGISYY770 pKa = 10.53SPSLVLLNAGDD781 pKa = 4.08GTFPTSITLPGGSPEE796 pKa = 4.28TNSIAAADD804 pKa = 3.73VDD806 pKa = 4.1GDD808 pKa = 3.71GDD810 pKa = 4.7LDD812 pKa = 3.91VLVGYY817 pKa = 9.93SLVLNTGNGTFPTRR831 pKa = 11.84ITLPGGSPATQSIAAADD848 pKa = 3.63VDD850 pKa = 4.1GDD852 pKa = 3.77GDD854 pKa = 5.12LDD856 pKa = 3.75VLLGNFGSPSQVLLNAGDD874 pKa = 4.07GTFPTSMEE882 pKa = 4.12LPGGPRR888 pKa = 11.84EE889 pKa = 4.29ATFSIAAADD898 pKa = 3.94LDD900 pKa = 4.16GDD902 pKa = 4.11GDD904 pKa = 4.47SDD906 pKa = 4.8LLLGNSGNPCRR917 pKa = 11.84VLLNAGSRR925 pKa = 11.84SGGITFEE932 pKa = 4.4TIIEE936 pKa = 4.44INTGTRR942 pKa = 11.84IYY944 pKa = 10.51EE945 pKa = 4.07PSPLTNSIVAADD957 pKa = 3.64VDD959 pKa = 4.17GDD961 pKa = 3.94GDD963 pKa = 5.55LDD965 pKa = 4.5LLLANSNNYY974 pKa = 8.84MNLKK978 pKa = 10.22SSLVLLNAGDD988 pKa = 4.02GTFPTIIKK996 pKa = 10.25LIGGSADD1003 pKa = 3.6TNSITDD1009 pKa = 3.52TSSIAAADD1017 pKa = 3.67VDD1019 pKa = 4.26SDD1021 pKa = 3.78GDD1023 pKa = 4.14LDD1025 pKa = 3.96VLLGGSPGHH1034 pKa = 6.88PSLVLLNAGNGTFPTSIEE1052 pKa = 4.09LPGGSEE1058 pKa = 4.14GTNSIAAADD1067 pKa = 3.73VDD1069 pKa = 4.1GDD1071 pKa = 3.77GDD1073 pKa = 4.97LDD1075 pKa = 3.83VLLGGNPSRR1084 pKa = 11.84VLLNAGGGTFPTSIEE1099 pKa = 4.16LPGSEE1104 pKa = 4.17GTNSIAAADD1113 pKa = 3.73VDD1115 pKa = 4.1GDD1117 pKa = 3.77GDD1119 pKa = 5.36LDD1121 pKa = 3.72VLLAGWFNRR1130 pKa = 11.84VLLNAGDD1137 pKa = 4.08GTFPASITLPSHH1149 pKa = 7.06DD1150 pKa = 3.5GSDD1153 pKa = 3.69RR1154 pKa = 11.84IVAADD1159 pKa = 4.07LDD1161 pKa = 4.37GDD1163 pKa = 3.96GDD1165 pKa = 4.99LDD1167 pKa = 3.75VLLGNRR1173 pKa = 11.84NSPSLVVLNAGNGTFPTSISLPGSSKK1199 pKa = 10.86LGISLAVADD1208 pKa = 4.51MDD1210 pKa = 4.8GDD1212 pKa = 3.98GDD1214 pKa = 4.79LDD1216 pKa = 3.95VLIGKK1221 pKa = 9.76YY1222 pKa = 10.23GLPSLLLLNAGDD1234 pKa = 4.11GTFPTSIEE1242 pKa = 4.14LPGGSAEE1249 pKa = 4.22TSSIAAADD1257 pKa = 3.55VDD1259 pKa = 4.1GDD1261 pKa = 3.74GDD1263 pKa = 4.8LDD1265 pKa = 3.82VLIGNRR1271 pKa = 11.84NSPSLVLPFIRR1282 pKa = 11.84CSDD1285 pKa = 3.58PGTARR1290 pKa = 11.84SRR1292 pKa = 11.84YY1293 pKa = 8.97GNGCVRR1299 pKa = 11.84CPAPSSRR1306 pKa = 11.84RR1307 pKa = 11.84DD1308 pKa = 3.64DD1309 pKa = 3.96VSDD1312 pKa = 3.41VCYY1315 pKa = 10.57EE1316 pKa = 4.07CDD1318 pKa = 3.37EE1319 pKa = 4.93HH1320 pKa = 8.75SHH1322 pKa = 6.93LDD1324 pKa = 3.74PGSGEE1329 pKa = 4.19CVACTEE1335 pKa = 4.16GSEE1338 pKa = 4.2RR1339 pKa = 11.84LLGATRR1345 pKa = 11.84CSRR1348 pKa = 11.84CPVGKK1353 pKa = 9.94RR1354 pKa = 11.84HH1355 pKa = 6.19AATGTGCIPCSPGKK1369 pKa = 8.93FTSTSGSIACQPCTPGYY1386 pKa = 8.81MCIEE1390 pKa = 4.81GSSAPQPCPGGTHH1403 pKa = 7.48ANQTMLNLTGYY1414 pKa = 10.14LGSLSEE1420 pKa = 4.89CLICPVGTSCSVGSAVAVPCAPGTYY1445 pKa = 9.84NNRR1448 pKa = 11.84SGQEE1452 pKa = 3.89TCLKK1456 pKa = 9.59CAPGSFQEE1464 pKa = 4.37LAGEE1468 pKa = 4.87TSCSPCPAGYY1478 pKa = 10.42YY1479 pKa = 9.07CAEE1482 pKa = 4.32GAAAALPCPFGTHH1495 pKa = 5.86KK1496 pKa = 10.68NASLSVMTSVDD1507 pKa = 3.3QCVVCPVGTSCSVGSAVAVPCAPGTYY1533 pKa = 9.94NNQPRR1538 pKa = 11.84QEE1540 pKa = 4.19TCLKK1544 pKa = 9.76CAAGSFQEE1552 pKa = 4.16LAGRR1556 pKa = 11.84TSCNPCTPGYY1566 pKa = 10.16YY1567 pKa = 9.44CAEE1570 pKa = 4.42GAATPVPCPRR1580 pKa = 11.84GKK1582 pKa = 10.47YY1583 pKa = 9.16NNRR1586 pKa = 11.84TGLEE1590 pKa = 4.04FAVGCVPVGFGFWAPTGSDD1609 pKa = 3.78FPIPCPSGFSCPGYY1623 pKa = 10.88FNDD1626 pKa = 4.04NLTFGSQPLILPSGGLPPEE1645 pKa = 4.4VAVEE1649 pKa = 4.19TQTVEE1654 pKa = 4.0TATGSISLNTPLSSFLNDD1672 pKa = 3.01TALQLQYY1679 pKa = 10.25RR1680 pKa = 11.84QTIAAQLGIPWQSLQLDD1697 pKa = 4.8FSAAPPSVGRR1707 pKa = 11.84RR1708 pKa = 11.84LQTSSTIVTYY1718 pKa = 9.41TIVRR1722 pKa = 11.84TTTTVHH1728 pKa = 6.43EE1729 pKa = 4.81GADD1732 pKa = 3.61SVSQCVCKK1740 pKa = 10.72LGTFASLTRR1749 pKa = 11.84AEE1751 pKa = 4.67MMQATNDD1758 pKa = 3.66SSRR1761 pKa = 11.84VAADD1765 pKa = 3.09AFEE1768 pKa = 4.01
MM1 pKa = 7.47SFADD5 pKa = 3.78AQQCATTCIGAPGYY19 pKa = 11.08ASDD22 pKa = 4.75CDD24 pKa = 3.94CAAEE28 pKa = 4.01FASNKK33 pKa = 9.44FATEE37 pKa = 3.7YY38 pKa = 10.52GYY40 pKa = 11.19DD41 pKa = 4.29FITVNGVSSSLTNGPMNVQVATEE64 pKa = 4.15TAQLVDD70 pKa = 3.8TFSTTTFGAVQACTRR85 pKa = 11.84GSLEE89 pKa = 3.96TFKK92 pKa = 11.29ARR94 pKa = 11.84VAAKK98 pKa = 9.84SLSNMTSTGAKK109 pKa = 8.93MDD111 pKa = 3.53PTLNDD116 pKa = 3.05VQTTLSIISYY126 pKa = 10.31LGGLLEE132 pKa = 4.29TDD134 pKa = 3.05AFEE137 pKa = 4.81ANSTIDD143 pKa = 3.12IAEE146 pKa = 4.56AAHH149 pKa = 5.93EE150 pKa = 4.26TDD152 pKa = 4.48PDD154 pKa = 3.99SMPYY158 pKa = 10.36ARR160 pKa = 11.84SSSTGASLVAIAPALLPGMLAVLGLLCAVLYY191 pKa = 10.47GAVQAFAAHH200 pKa = 6.94LGASALASIMGPPPPKK216 pKa = 10.08GRR218 pKa = 11.84RR219 pKa = 11.84AVSAAPASCRR229 pKa = 11.84PSARR233 pKa = 11.84RR234 pKa = 11.84GSAGRR239 pKa = 11.84FGSAPRR245 pKa = 11.84LVGFLLLALFVSATDD260 pKa = 3.77AQCMEE265 pKa = 4.58EE266 pKa = 4.01WCNVCTGEE274 pKa = 4.4CTSATGGEE282 pKa = 4.18LLGSSEE288 pKa = 4.94AVLQFLSSVLPSSSSGLTSSTTFSIATADD317 pKa = 3.39VDD319 pKa = 4.08GDD321 pKa = 3.75GDD323 pKa = 4.97LDD325 pKa = 3.87VLLGGSPTSRR335 pKa = 11.84VLLNAGNGTFPTSIEE350 pKa = 4.1LPGVSAEE357 pKa = 4.19TRR359 pKa = 11.84SIAAADD365 pKa = 3.88LDD367 pKa = 4.21GDD369 pKa = 3.95GDD371 pKa = 4.83LDD373 pKa = 3.87VLLGSSPSSVLLNAGDD389 pKa = 4.03GTFPTSITLPGDD401 pKa = 3.58GALTRR406 pKa = 11.84SITAADD412 pKa = 4.01LDD414 pKa = 4.23GDD416 pKa = 3.94GDD418 pKa = 4.83LDD420 pKa = 3.93VLIGNSGSPSRR431 pKa = 11.84VLLNAGDD438 pKa = 4.08GTFPASIEE446 pKa = 4.12LPGGSPLTYY455 pKa = 10.15SIAAADD461 pKa = 3.63VDD463 pKa = 4.1GDD465 pKa = 3.74GDD467 pKa = 4.8LDD469 pKa = 3.92VLIGNSNSPNRR480 pKa = 11.84VLLNAGDD487 pKa = 4.09GTFPTSITLPGGSAEE502 pKa = 4.36TNSIAAADD510 pKa = 3.73VDD512 pKa = 4.1GDD514 pKa = 3.74GDD516 pKa = 4.8LDD518 pKa = 3.92VLIGNSNSPNRR529 pKa = 11.84VLLNTGNGTFPTSIEE544 pKa = 4.17LPGSIAATQSIAAADD559 pKa = 3.63VDD561 pKa = 4.1GDD563 pKa = 3.77GDD565 pKa = 5.12LDD567 pKa = 3.87VLLGNNGSPSRR578 pKa = 11.84VLLNAGNGTFPTSITLPGGSLFTQSIAAADD608 pKa = 3.63VDD610 pKa = 4.1GDD612 pKa = 3.74GDD614 pKa = 4.8LDD616 pKa = 3.87VLIGNSQYY624 pKa = 10.96RR625 pKa = 11.84NRR627 pKa = 11.84MLLNAGGTFQTIIQLPGPGIVSTHH651 pKa = 6.58SIAAADD657 pKa = 4.29LDD659 pKa = 4.12SDD661 pKa = 3.96GDD663 pKa = 4.15LDD665 pKa = 4.27VLIGNSGSPSQVLLNAGDD683 pKa = 3.99GTFPASITLPGGSPFTQSVAAADD706 pKa = 3.67VDD708 pKa = 3.96GDD710 pKa = 3.74GDD712 pKa = 4.8LDD714 pKa = 3.85VLIGNSYY721 pKa = 10.75SPSLVLLNAGDD732 pKa = 4.08GTFPTSIEE740 pKa = 4.19LPGGTGDD747 pKa = 3.5TRR749 pKa = 11.84SIAAADD755 pKa = 3.54VDD757 pKa = 4.1GDD759 pKa = 3.77GDD761 pKa = 4.83LDD763 pKa = 3.79VLLGISYY770 pKa = 10.53SPSLVLLNAGDD781 pKa = 4.08GTFPTSITLPGGSPEE796 pKa = 4.28TNSIAAADD804 pKa = 3.73VDD806 pKa = 4.1GDD808 pKa = 3.71GDD810 pKa = 4.7LDD812 pKa = 3.91VLVGYY817 pKa = 9.93SLVLNTGNGTFPTRR831 pKa = 11.84ITLPGGSPATQSIAAADD848 pKa = 3.63VDD850 pKa = 4.1GDD852 pKa = 3.77GDD854 pKa = 5.12LDD856 pKa = 3.75VLLGNFGSPSQVLLNAGDD874 pKa = 4.07GTFPTSMEE882 pKa = 4.12LPGGPRR888 pKa = 11.84EE889 pKa = 4.29ATFSIAAADD898 pKa = 3.94LDD900 pKa = 4.16GDD902 pKa = 4.11GDD904 pKa = 4.47SDD906 pKa = 4.8LLLGNSGNPCRR917 pKa = 11.84VLLNAGSRR925 pKa = 11.84SGGITFEE932 pKa = 4.4TIIEE936 pKa = 4.44INTGTRR942 pKa = 11.84IYY944 pKa = 10.51EE945 pKa = 4.07PSPLTNSIVAADD957 pKa = 3.64VDD959 pKa = 4.17GDD961 pKa = 3.94GDD963 pKa = 5.55LDD965 pKa = 4.5LLLANSNNYY974 pKa = 8.84MNLKK978 pKa = 10.22SSLVLLNAGDD988 pKa = 4.02GTFPTIIKK996 pKa = 10.25LIGGSADD1003 pKa = 3.6TNSITDD1009 pKa = 3.52TSSIAAADD1017 pKa = 3.67VDD1019 pKa = 4.26SDD1021 pKa = 3.78GDD1023 pKa = 4.14LDD1025 pKa = 3.96VLLGGSPGHH1034 pKa = 6.88PSLVLLNAGNGTFPTSIEE1052 pKa = 4.09LPGGSEE1058 pKa = 4.14GTNSIAAADD1067 pKa = 3.73VDD1069 pKa = 4.1GDD1071 pKa = 3.77GDD1073 pKa = 4.97LDD1075 pKa = 3.83VLLGGNPSRR1084 pKa = 11.84VLLNAGGGTFPTSIEE1099 pKa = 4.16LPGSEE1104 pKa = 4.17GTNSIAAADD1113 pKa = 3.73VDD1115 pKa = 4.1GDD1117 pKa = 3.77GDD1119 pKa = 5.36LDD1121 pKa = 3.72VLLAGWFNRR1130 pKa = 11.84VLLNAGDD1137 pKa = 4.08GTFPASITLPSHH1149 pKa = 7.06DD1150 pKa = 3.5GSDD1153 pKa = 3.69RR1154 pKa = 11.84IVAADD1159 pKa = 4.07LDD1161 pKa = 4.37GDD1163 pKa = 3.96GDD1165 pKa = 4.99LDD1167 pKa = 3.75VLLGNRR1173 pKa = 11.84NSPSLVVLNAGNGTFPTSISLPGSSKK1199 pKa = 10.86LGISLAVADD1208 pKa = 4.51MDD1210 pKa = 4.8GDD1212 pKa = 3.98GDD1214 pKa = 4.79LDD1216 pKa = 3.95VLIGKK1221 pKa = 9.76YY1222 pKa = 10.23GLPSLLLLNAGDD1234 pKa = 4.11GTFPTSIEE1242 pKa = 4.14LPGGSAEE1249 pKa = 4.22TSSIAAADD1257 pKa = 3.55VDD1259 pKa = 4.1GDD1261 pKa = 3.74GDD1263 pKa = 4.8LDD1265 pKa = 3.82VLIGNRR1271 pKa = 11.84NSPSLVLPFIRR1282 pKa = 11.84CSDD1285 pKa = 3.58PGTARR1290 pKa = 11.84SRR1292 pKa = 11.84YY1293 pKa = 8.97GNGCVRR1299 pKa = 11.84CPAPSSRR1306 pKa = 11.84RR1307 pKa = 11.84DD1308 pKa = 3.64DD1309 pKa = 3.96VSDD1312 pKa = 3.41VCYY1315 pKa = 10.57EE1316 pKa = 4.07CDD1318 pKa = 3.37EE1319 pKa = 4.93HH1320 pKa = 8.75SHH1322 pKa = 6.93LDD1324 pKa = 3.74PGSGEE1329 pKa = 4.19CVACTEE1335 pKa = 4.16GSEE1338 pKa = 4.2RR1339 pKa = 11.84LLGATRR1345 pKa = 11.84CSRR1348 pKa = 11.84CPVGKK1353 pKa = 9.94RR1354 pKa = 11.84HH1355 pKa = 6.19AATGTGCIPCSPGKK1369 pKa = 8.93FTSTSGSIACQPCTPGYY1386 pKa = 8.81MCIEE1390 pKa = 4.81GSSAPQPCPGGTHH1403 pKa = 7.48ANQTMLNLTGYY1414 pKa = 10.14LGSLSEE1420 pKa = 4.89CLICPVGTSCSVGSAVAVPCAPGTYY1445 pKa = 9.84NNRR1448 pKa = 11.84SGQEE1452 pKa = 3.89TCLKK1456 pKa = 9.59CAPGSFQEE1464 pKa = 4.37LAGEE1468 pKa = 4.87TSCSPCPAGYY1478 pKa = 10.42YY1479 pKa = 9.07CAEE1482 pKa = 4.32GAAAALPCPFGTHH1495 pKa = 5.86KK1496 pKa = 10.68NASLSVMTSVDD1507 pKa = 3.3QCVVCPVGTSCSVGSAVAVPCAPGTYY1533 pKa = 9.94NNQPRR1538 pKa = 11.84QEE1540 pKa = 4.19TCLKK1544 pKa = 9.76CAAGSFQEE1552 pKa = 4.16LAGRR1556 pKa = 11.84TSCNPCTPGYY1566 pKa = 10.16YY1567 pKa = 9.44CAEE1570 pKa = 4.42GAATPVPCPRR1580 pKa = 11.84GKK1582 pKa = 10.47YY1583 pKa = 9.16NNRR1586 pKa = 11.84TGLEE1590 pKa = 4.04FAVGCVPVGFGFWAPTGSDD1609 pKa = 3.78FPIPCPSGFSCPGYY1623 pKa = 10.88FNDD1626 pKa = 4.04NLTFGSQPLILPSGGLPPEE1645 pKa = 4.4VAVEE1649 pKa = 4.19TQTVEE1654 pKa = 4.0TATGSISLNTPLSSFLNDD1672 pKa = 3.01TALQLQYY1679 pKa = 10.25RR1680 pKa = 11.84QTIAAQLGIPWQSLQLDD1697 pKa = 4.8FSAAPPSVGRR1707 pKa = 11.84RR1708 pKa = 11.84LQTSSTIVTYY1718 pKa = 9.41TIVRR1722 pKa = 11.84TTTTVHH1728 pKa = 6.43EE1729 pKa = 4.81GADD1732 pKa = 3.61SVSQCVCKK1740 pKa = 10.72LGTFASLTRR1749 pKa = 11.84AEE1751 pKa = 4.67MMQATNDD1758 pKa = 3.66SSRR1761 pKa = 11.84VAADD1765 pKa = 3.09AFEE1768 pKa = 4.01
Molecular weight: 177.92 kDa
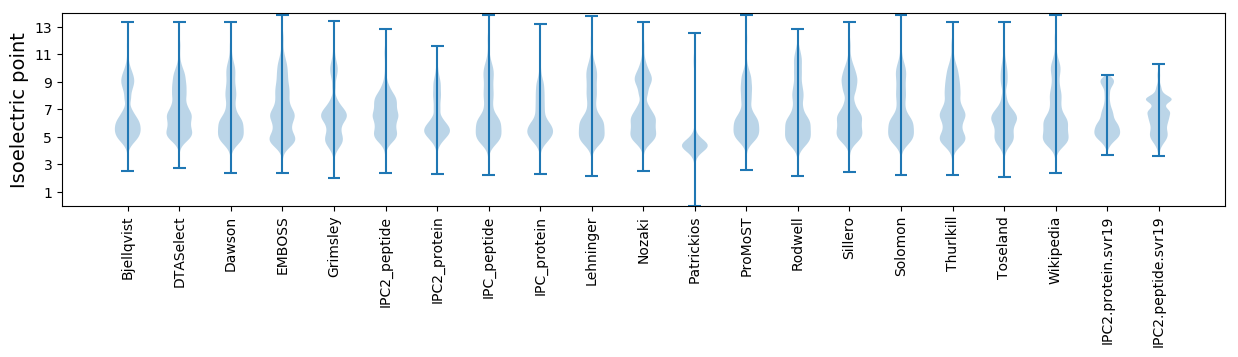
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M0J5Z7|A0A0M0J5Z7_9EUKA Membrane protein OS=Chrysochromulina tobinii OX=1460289 GN=Ctob_000966 PE=4 SV=1
MM1 pKa = 7.7PRR3 pKa = 11.84TSPEE7 pKa = 3.73PAQATAASRR16 pKa = 11.84SSSGGSPLPSMQLRR30 pKa = 11.84RR31 pKa = 11.84AALARR36 pKa = 11.84PPRR39 pKa = 11.84NSRR42 pKa = 11.84SRR44 pKa = 11.84LGVSRR49 pKa = 11.84STSRR53 pKa = 11.84TAATLTVKK61 pKa = 10.5PSQRR65 pKa = 11.84SRR67 pKa = 11.84ARR69 pKa = 11.84LGSILPAAAASHH81 pKa = 5.94GRR83 pKa = 11.84PASTRR88 pKa = 11.84TLMLRR93 pKa = 11.84SVISASSVGAGG104 pKa = 3.2
MM1 pKa = 7.7PRR3 pKa = 11.84TSPEE7 pKa = 3.73PAQATAASRR16 pKa = 11.84SSSGGSPLPSMQLRR30 pKa = 11.84RR31 pKa = 11.84AALARR36 pKa = 11.84PPRR39 pKa = 11.84NSRR42 pKa = 11.84SRR44 pKa = 11.84LGVSRR49 pKa = 11.84STSRR53 pKa = 11.84TAATLTVKK61 pKa = 10.5PSQRR65 pKa = 11.84SRR67 pKa = 11.84ARR69 pKa = 11.84LGSILPAAAASHH81 pKa = 5.94GRR83 pKa = 11.84PASTRR88 pKa = 11.84TLMLRR93 pKa = 11.84SVISASSVGAGG104 pKa = 3.2
Molecular weight: 10.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
7768616 |
10 |
10901 |
465.8 |
50.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.134 ± 0.042 | 1.573 ± 0.009 |
5.19 ± 0.013 | 6.665 ± 0.025 |
3.082 ± 0.013 | 7.575 ± 0.023 |
2.29 ± 0.009 | 3.438 ± 0.014 |
4.125 ± 0.025 | 10.169 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.357 ± 0.009 | 2.423 ± 0.011 |
5.47 ± 0.018 | 3.529 ± 0.013 |
6.754 ± 0.023 | 6.892 ± 0.017 |
5.227 ± 0.014 | 6.706 ± 0.014 |
1.336 ± 0.008 | 2.066 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
