
Arthrobacter sp. Edens01
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Arthrobacter; unclassified Arthrobacter
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
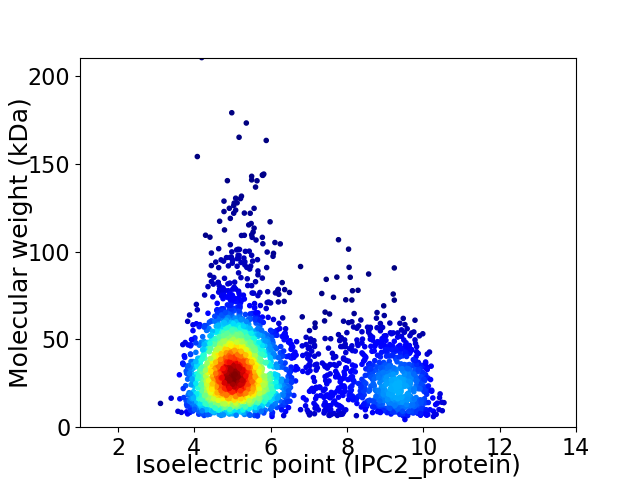
Virtual 2D-PAGE plot for 3212 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N8HX36|A0A0N8HX36_9MICC Uncharacterized protein OS=Arthrobacter sp. Edens01 OX=1732020 GN=AO716_16090 PE=3 SV=1
MM1 pKa = 6.85TRR3 pKa = 11.84ALIVVDD9 pKa = 3.88VQNDD13 pKa = 3.59FCEE16 pKa = 4.95GGSLAVPGGNDD27 pKa = 3.02TAAEE31 pKa = 3.81ISDD34 pKa = 4.26YY35 pKa = 11.21IEE37 pKa = 4.01TSRR40 pKa = 11.84DD41 pKa = 2.95RR42 pKa = 11.84YY43 pKa = 10.61AVVAATQDD51 pKa = 3.1WHH53 pKa = 7.42IEE55 pKa = 4.13PGSHH59 pKa = 7.5FSDD62 pKa = 3.25NPNFRR67 pKa = 11.84DD68 pKa = 3.44SWPVHH73 pKa = 5.95CVAGTHH79 pKa = 6.15GAALNPDD86 pKa = 4.32LDD88 pKa = 4.05TEE90 pKa = 4.82FIDD93 pKa = 4.35AYY95 pKa = 10.2FRR97 pKa = 11.84KK98 pKa = 9.54GQYY101 pKa = 8.2EE102 pKa = 3.74AAYY105 pKa = 10.48SGFEE109 pKa = 4.04GLLAPDD115 pKa = 3.74VEE117 pKa = 4.88VPLGDD122 pKa = 4.26PDD124 pKa = 5.11AEE126 pKa = 4.3PEE128 pKa = 4.02PDD130 pKa = 3.53EE131 pKa = 5.48DD132 pKa = 5.71AVSLDD137 pKa = 3.07DD138 pKa = 3.68WLRR141 pKa = 11.84DD142 pKa = 3.58NGVDD146 pKa = 3.34EE147 pKa = 4.33VVIVGLATDD156 pKa = 3.84YY157 pKa = 10.85CVKK160 pKa = 10.11ATALDD165 pKa = 3.86AVAAGYY171 pKa = 7.79ATYY174 pKa = 10.5VIPEE178 pKa = 4.64LCRR181 pKa = 11.84GLDD184 pKa = 3.53KK185 pKa = 11.37VDD187 pKa = 3.45VLAALDD193 pKa = 3.9EE194 pKa = 4.8LEE196 pKa = 4.48DD197 pKa = 4.47AGVEE201 pKa = 4.39LLDD204 pKa = 4.28LL205 pKa = 4.67
MM1 pKa = 6.85TRR3 pKa = 11.84ALIVVDD9 pKa = 3.88VQNDD13 pKa = 3.59FCEE16 pKa = 4.95GGSLAVPGGNDD27 pKa = 3.02TAAEE31 pKa = 3.81ISDD34 pKa = 4.26YY35 pKa = 11.21IEE37 pKa = 4.01TSRR40 pKa = 11.84DD41 pKa = 2.95RR42 pKa = 11.84YY43 pKa = 10.61AVVAATQDD51 pKa = 3.1WHH53 pKa = 7.42IEE55 pKa = 4.13PGSHH59 pKa = 7.5FSDD62 pKa = 3.25NPNFRR67 pKa = 11.84DD68 pKa = 3.44SWPVHH73 pKa = 5.95CVAGTHH79 pKa = 6.15GAALNPDD86 pKa = 4.32LDD88 pKa = 4.05TEE90 pKa = 4.82FIDD93 pKa = 4.35AYY95 pKa = 10.2FRR97 pKa = 11.84KK98 pKa = 9.54GQYY101 pKa = 8.2EE102 pKa = 3.74AAYY105 pKa = 10.48SGFEE109 pKa = 4.04GLLAPDD115 pKa = 3.74VEE117 pKa = 4.88VPLGDD122 pKa = 4.26PDD124 pKa = 5.11AEE126 pKa = 4.3PEE128 pKa = 4.02PDD130 pKa = 3.53EE131 pKa = 5.48DD132 pKa = 5.71AVSLDD137 pKa = 3.07DD138 pKa = 3.68WLRR141 pKa = 11.84DD142 pKa = 3.58NGVDD146 pKa = 3.34EE147 pKa = 4.33VVIVGLATDD156 pKa = 3.84YY157 pKa = 10.85CVKK160 pKa = 10.11ATALDD165 pKa = 3.86AVAAGYY171 pKa = 7.79ATYY174 pKa = 10.5VIPEE178 pKa = 4.64LCRR181 pKa = 11.84GLDD184 pKa = 3.53KK185 pKa = 11.37VDD187 pKa = 3.45VLAALDD193 pKa = 3.9EE194 pKa = 4.8LEE196 pKa = 4.48DD197 pKa = 4.47AGVEE201 pKa = 4.39LLDD204 pKa = 4.28LL205 pKa = 4.67
Molecular weight: 22.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P7G887|A0A0P7G887_9MICC Uncharacterized protein OS=Arthrobacter sp. Edens01 OX=1732020 GN=AO716_08540 PE=4 SV=1
MM1 pKa = 7.28TGIAIACALASAVFLAFGAQRR22 pKa = 11.84QGSAVSADD30 pKa = 3.4TGGLSLNSAGLGRR43 pKa = 11.84LLRR46 pKa = 11.84NPRR49 pKa = 11.84WLFGLLLLGVGTGLNVAALAMAPLTVVQPIGSLALVITTIVNSRR93 pKa = 11.84DD94 pKa = 2.87QGLRR98 pKa = 11.84LNRR101 pKa = 11.84VTVVSIVACVLGSMLFVLLAVGATRR126 pKa = 11.84SEE128 pKa = 4.27PVVEE132 pKa = 4.05ARR134 pKa = 11.84QEE136 pKa = 3.92IVIVLILAVVVAFFGMLNLVFGKK159 pKa = 10.41RR160 pKa = 11.84LGAIAHH166 pKa = 6.48ILGAGILFGFVAVLTKK182 pKa = 10.19TIAADD187 pKa = 3.66LLDD190 pKa = 4.26PNGRR194 pKa = 11.84FLLNVPWYY202 pKa = 8.66TIVGIAVAGGLGAWFVQSAYY222 pKa = 11.06SSGPPDD228 pKa = 3.76LVIAGLTVIDD238 pKa = 4.44PMVGIAIGIGVLNEE252 pKa = 4.06LRR254 pKa = 11.84PDD256 pKa = 3.39VPAVLGVAMGVCAVIAIVGVVALSRR281 pKa = 11.84YY282 pKa = 9.23HH283 pKa = 6.81PDD285 pKa = 3.24VIKK288 pKa = 10.58RR289 pKa = 11.84RR290 pKa = 11.84AMNKK294 pKa = 8.92RR295 pKa = 11.84RR296 pKa = 11.84QKK298 pKa = 10.21PVSTRR303 pKa = 11.84ATPTNKK309 pKa = 9.81PP310 pKa = 3.25
MM1 pKa = 7.28TGIAIACALASAVFLAFGAQRR22 pKa = 11.84QGSAVSADD30 pKa = 3.4TGGLSLNSAGLGRR43 pKa = 11.84LLRR46 pKa = 11.84NPRR49 pKa = 11.84WLFGLLLLGVGTGLNVAALAMAPLTVVQPIGSLALVITTIVNSRR93 pKa = 11.84DD94 pKa = 2.87QGLRR98 pKa = 11.84LNRR101 pKa = 11.84VTVVSIVACVLGSMLFVLLAVGATRR126 pKa = 11.84SEE128 pKa = 4.27PVVEE132 pKa = 4.05ARR134 pKa = 11.84QEE136 pKa = 3.92IVIVLILAVVVAFFGMLNLVFGKK159 pKa = 10.41RR160 pKa = 11.84LGAIAHH166 pKa = 6.48ILGAGILFGFVAVLTKK182 pKa = 10.19TIAADD187 pKa = 3.66LLDD190 pKa = 4.26PNGRR194 pKa = 11.84FLLNVPWYY202 pKa = 8.66TIVGIAVAGGLGAWFVQSAYY222 pKa = 11.06SSGPPDD228 pKa = 3.76LVIAGLTVIDD238 pKa = 4.44PMVGIAIGIGVLNEE252 pKa = 4.06LRR254 pKa = 11.84PDD256 pKa = 3.39VPAVLGVAMGVCAVIAIVGVVALSRR281 pKa = 11.84YY282 pKa = 9.23HH283 pKa = 6.81PDD285 pKa = 3.24VIKK288 pKa = 10.58RR289 pKa = 11.84RR290 pKa = 11.84AMNKK294 pKa = 8.92RR295 pKa = 11.84RR296 pKa = 11.84QKK298 pKa = 10.21PVSTRR303 pKa = 11.84ATPTNKK309 pKa = 9.81PP310 pKa = 3.25
Molecular weight: 32.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1044346 |
37 |
2033 |
325.1 |
34.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.16 ± 0.073 | 0.58 ± 0.009 |
5.601 ± 0.037 | 6.042 ± 0.041 |
3.195 ± 0.03 | 9.119 ± 0.039 |
1.874 ± 0.02 | 4.22 ± 0.033 |
2.466 ± 0.032 | 10.483 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.035 ± 0.017 | 2.399 ± 0.025 |
5.518 ± 0.033 | 3.114 ± 0.023 |
6.704 ± 0.038 | 5.963 ± 0.031 |
5.702 ± 0.031 | 8.267 ± 0.034 |
1.426 ± 0.02 | 2.134 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
