
Ferret coronavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Minacovirus
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
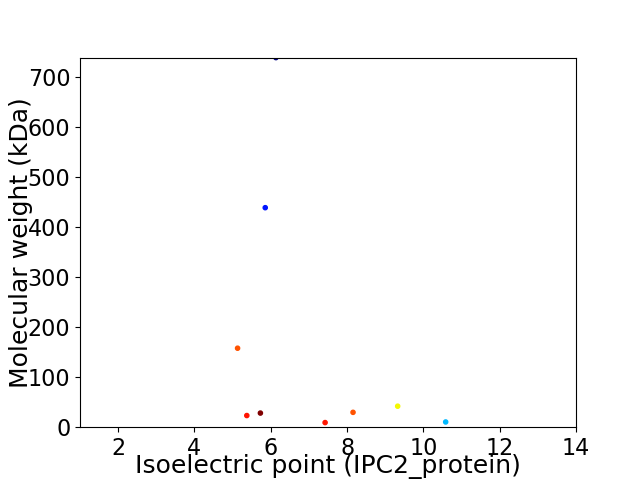
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A172B201|A0A172B201_9ALPC 3C-like proteinase OS=Ferret coronavirus OX=1264898 GN=orf1ab PE=3 SV=1
MM1 pKa = 7.76FEE3 pKa = 3.53MRR5 pKa = 11.84TPVFLTVCIVLLQYY19 pKa = 10.04AHH21 pKa = 7.7CDD23 pKa = 3.25TFNCSTLHH31 pKa = 6.06NTTLWATDD39 pKa = 3.21QTRR42 pKa = 11.84LLDD45 pKa = 3.83YY46 pKa = 10.4FIANYY51 pKa = 9.97SSRR54 pKa = 11.84LPTGASVVLGDD65 pKa = 4.08YY66 pKa = 10.67FPTLGPWYY74 pKa = 9.74DD75 pKa = 3.74CVSNTTYY82 pKa = 10.99GGVVLEE88 pKa = 4.91DD89 pKa = 3.64LRR91 pKa = 11.84ALYY94 pKa = 10.36LDD96 pKa = 3.87HH97 pKa = 6.27QQGVARR103 pKa = 11.84DD104 pKa = 3.56LAFAVYY110 pKa = 9.27GYY112 pKa = 9.21PYY114 pKa = 9.57TAIVVLNKK122 pKa = 10.26EE123 pKa = 4.23LFNGQGYY130 pKa = 10.42GGLLCFCNGNSTIPAFNANCSTSCTVRR157 pKa = 11.84SFRR160 pKa = 11.84LCADD164 pKa = 3.34EE165 pKa = 5.13SICHH169 pKa = 5.41EE170 pKa = 5.0RR171 pKa = 11.84ILGLKK176 pKa = 8.26WSSSEE181 pKa = 3.92VVAYY185 pKa = 10.14LAGEE189 pKa = 4.35VYY191 pKa = 10.35SYY193 pKa = 11.35KK194 pKa = 10.73LSNHH198 pKa = 6.35WYY200 pKa = 9.45NNVTIKK206 pKa = 9.41TQTNANQIYY215 pKa = 8.12WWFNPVRR222 pKa = 11.84DD223 pKa = 3.67LSYY226 pKa = 10.96YY227 pKa = 10.35NVNKK231 pKa = 9.63TVDD234 pKa = 3.4STIVVSNCTSDD245 pKa = 3.43CSGYY249 pKa = 10.39AANIFAVEE257 pKa = 4.12TGGFIPSSFSFNNWFVLTNSSTIVSGKK284 pKa = 9.78FVSSQPLLVNCLTPVPSFGDD304 pKa = 3.31EE305 pKa = 3.8TSIINFDD312 pKa = 4.06TVPSQCNGATVNGSFDD328 pKa = 3.49VVRR331 pKa = 11.84FNLNFTANVASSSGTSFLALNTTGGVVLYY360 pKa = 8.29LSCFNEE366 pKa = 4.25TKK368 pKa = 8.19TTNLYY373 pKa = 9.43MSEE376 pKa = 4.06GALPFGTHH384 pKa = 6.29EE385 pKa = 4.34GALYY389 pKa = 10.53CYY391 pKa = 10.38VSYY394 pKa = 11.56NEE396 pKa = 4.29TSFKK400 pKa = 10.75YY401 pKa = 10.14IGVLPPSVKK410 pKa = 10.29EE411 pKa = 3.65IAVSKK416 pKa = 10.09WGFVYY421 pKa = 10.59INGYY425 pKa = 10.63NYY427 pKa = 10.16FQTFPIDD434 pKa = 3.43SVAFNLTTGNSGAFWTIAYY453 pKa = 7.01TTFTDD458 pKa = 3.4VLLEE462 pKa = 4.21VSDD465 pKa = 3.99TQIKK469 pKa = 10.22SVTYY473 pKa = 10.18CNSHH477 pKa = 6.99INDD480 pKa = 3.81IKK482 pKa = 10.42CSQMSEE488 pKa = 4.0NLPDD492 pKa = 3.23GFYY495 pKa = 10.52PVSQHH500 pKa = 6.53SLPNVNKK507 pKa = 9.6TFVTLPANFEE517 pKa = 4.14HH518 pKa = 6.76TSINVTGNVLLIYY531 pKa = 9.48YY532 pKa = 7.1AHH534 pKa = 6.84PVFRR538 pKa = 11.84SGNVTLHH545 pKa = 5.83PQGTQTICVNTTQFTFNFDD564 pKa = 4.0SQCHH568 pKa = 4.18QLAGGGCDD576 pKa = 3.81GIGASMISIDD586 pKa = 4.92SGNCPFSPDD595 pKa = 4.12KK596 pKa = 11.18LNQHH600 pKa = 6.75LAFEE604 pKa = 4.49TLCFSTLPTGSDD616 pKa = 3.54CAFSLTVQNRR626 pKa = 11.84YY627 pKa = 5.17FTRR630 pKa = 11.84VFAHH634 pKa = 6.68LFVTYY639 pKa = 10.22KK640 pKa = 10.79YY641 pKa = 10.67GLDD644 pKa = 3.38HH645 pKa = 7.22LGVQTPDD652 pKa = 3.11VGVKK656 pKa = 10.15DD657 pKa = 4.02LSVVYY662 pKa = 9.85QNVCTEE668 pKa = 3.84YY669 pKa = 10.84NIYY672 pKa = 10.17GHH674 pKa = 7.04AGVGIIRR681 pKa = 11.84STNQTLLGGLYY692 pKa = 8.33YY693 pKa = 10.54TSLSGDD699 pKa = 3.35LLGFKK704 pKa = 10.35NVTTGEE710 pKa = 4.27VYY712 pKa = 10.74SIVPCQLSAQAAVINGKK729 pKa = 9.35IVGAVTSVQSPILDD743 pKa = 3.96LPHH746 pKa = 7.16HH747 pKa = 6.41IVTPQFYY754 pKa = 9.64YY755 pKa = 10.62HH756 pKa = 7.09SIYY759 pKa = 10.65NYY761 pKa = 9.87SATPTSYY768 pKa = 9.3RR769 pKa = 11.84TNNGFDD775 pKa = 3.47KK776 pKa = 11.17YY777 pKa = 10.98LVNCTPIITYY787 pKa = 10.93SNMGVCEE794 pKa = 3.87NGALVFINITQSEE807 pKa = 4.64NPVQPISTGNVTIPSNFTISVQVEE831 pKa = 3.96YY832 pKa = 11.03LQMSSEE838 pKa = 4.25AVSIDD843 pKa = 3.18CAQYY847 pKa = 10.24VCNGNPRR854 pKa = 11.84CNRR857 pKa = 11.84LLAQYY862 pKa = 10.42ISACHH867 pKa = 6.55AIEE870 pKa = 3.9QALITSTRR878 pKa = 11.84LEE880 pKa = 4.05ALEE883 pKa = 4.53LEE885 pKa = 4.52SMISISDD892 pKa = 3.57NALALATVEE901 pKa = 4.52TFNSSDD907 pKa = 3.63YY908 pKa = 11.15LDD910 pKa = 3.95PVYY913 pKa = 10.96NDD915 pKa = 3.1QHH917 pKa = 5.3NTIGGIYY924 pKa = 9.08MDD926 pKa = 4.39GLKK929 pKa = 10.72DD930 pKa = 3.47LLPRR934 pKa = 11.84RR935 pKa = 11.84SCANKK940 pKa = 9.99HH941 pKa = 4.29GTCRR945 pKa = 11.84SVIEE949 pKa = 3.89EE950 pKa = 4.11LLFNKK955 pKa = 9.48VVTSGLGTVDD965 pKa = 3.79EE966 pKa = 4.9DD967 pKa = 4.87YY968 pKa = 11.24KK969 pKa = 11.15RR970 pKa = 11.84CTNGLDD976 pKa = 3.62IADD979 pKa = 4.62LVCAQYY985 pKa = 11.58YY986 pKa = 8.99NGIMVLPGVVNADD999 pKa = 3.0KK1000 pKa = 10.45MAMYY1004 pKa = 8.05TASLAGGITLGALGGGLVSVPFATAVQARR1033 pKa = 11.84LNYY1036 pKa = 9.72VALQTDD1042 pKa = 4.22VLQQNQKK1049 pKa = 9.71ILAASFNQAIGNITLAFGKK1068 pKa = 10.32VNSAIQQTAQGLSTVAQALTKK1089 pKa = 10.33VQDD1092 pKa = 3.72VVNSQGKK1099 pKa = 9.12ALNHH1103 pKa = 5.94LTAQLQNNFQAISSSIEE1120 pKa = 3.99DD1121 pKa = 3.12IYY1123 pKa = 11.76YY1124 pKa = 10.95KK1125 pKa = 10.47LDD1127 pKa = 3.56EE1128 pKa = 4.62VNADD1132 pKa = 3.46AQVDD1136 pKa = 3.81RR1137 pKa = 11.84LITGRR1142 pKa = 11.84LASLNAFVTQTLTNQAQIRR1161 pKa = 11.84ASRR1164 pKa = 11.84QLSKK1168 pKa = 11.24EE1169 pKa = 4.03KK1170 pKa = 10.5INEE1173 pKa = 4.27CVRR1176 pKa = 11.84SQSSRR1181 pKa = 11.84FGFCGNGTHH1190 pKa = 7.49LFSLANAAPRR1200 pKa = 11.84GVMLFHH1206 pKa = 6.51TVLLPTSYY1214 pKa = 9.07KK1215 pKa = 10.18TVTAWSGVCAISNNKK1230 pKa = 8.2TMGFIVKK1237 pKa = 9.91DD1238 pKa = 3.55VSLTLFKK1245 pKa = 11.26NHH1247 pKa = 7.23DD1248 pKa = 3.99DD1249 pKa = 3.9KK1250 pKa = 11.61FYY1252 pKa = 9.47LTPRR1256 pKa = 11.84TMYY1259 pKa = 9.32EE1260 pKa = 3.65PRR1262 pKa = 11.84VATMSDD1268 pKa = 3.53FVQIEE1273 pKa = 4.54SCTTTFVNATVAEE1286 pKa = 4.51LPSIIPDD1293 pKa = 3.78YY1294 pKa = 11.1IDD1296 pKa = 3.31INGTIKK1302 pKa = 11.04DD1303 pKa = 3.94MLDD1306 pKa = 3.04QYY1308 pKa = 10.89KK1309 pKa = 10.44PNWTVPEE1316 pKa = 3.94LTIDD1320 pKa = 3.94VFNQTYY1326 pKa = 10.97LNLTGEE1332 pKa = 4.32INEE1335 pKa = 4.23LEE1337 pKa = 4.13NRR1339 pKa = 11.84SVILQQTVLEE1349 pKa = 4.5LEE1351 pKa = 4.27SLIDD1355 pKa = 4.28NINGTLVNLEE1365 pKa = 3.71WLNRR1369 pKa = 11.84VEE1371 pKa = 5.64TFVKK1375 pKa = 8.87WPWWVWVIIGLILLIALPMLLFCCLSTGCCGCCGCLTSCLAGCCKK1420 pKa = 10.33NSCKK1424 pKa = 10.19RR1425 pKa = 11.84PSYY1428 pKa = 9.43YY1429 pKa = 10.55EE1430 pKa = 3.75PMEE1433 pKa = 4.32KK1434 pKa = 10.25VHH1436 pKa = 7.08INN1438 pKa = 3.15
MM1 pKa = 7.76FEE3 pKa = 3.53MRR5 pKa = 11.84TPVFLTVCIVLLQYY19 pKa = 10.04AHH21 pKa = 7.7CDD23 pKa = 3.25TFNCSTLHH31 pKa = 6.06NTTLWATDD39 pKa = 3.21QTRR42 pKa = 11.84LLDD45 pKa = 3.83YY46 pKa = 10.4FIANYY51 pKa = 9.97SSRR54 pKa = 11.84LPTGASVVLGDD65 pKa = 4.08YY66 pKa = 10.67FPTLGPWYY74 pKa = 9.74DD75 pKa = 3.74CVSNTTYY82 pKa = 10.99GGVVLEE88 pKa = 4.91DD89 pKa = 3.64LRR91 pKa = 11.84ALYY94 pKa = 10.36LDD96 pKa = 3.87HH97 pKa = 6.27QQGVARR103 pKa = 11.84DD104 pKa = 3.56LAFAVYY110 pKa = 9.27GYY112 pKa = 9.21PYY114 pKa = 9.57TAIVVLNKK122 pKa = 10.26EE123 pKa = 4.23LFNGQGYY130 pKa = 10.42GGLLCFCNGNSTIPAFNANCSTSCTVRR157 pKa = 11.84SFRR160 pKa = 11.84LCADD164 pKa = 3.34EE165 pKa = 5.13SICHH169 pKa = 5.41EE170 pKa = 5.0RR171 pKa = 11.84ILGLKK176 pKa = 8.26WSSSEE181 pKa = 3.92VVAYY185 pKa = 10.14LAGEE189 pKa = 4.35VYY191 pKa = 10.35SYY193 pKa = 11.35KK194 pKa = 10.73LSNHH198 pKa = 6.35WYY200 pKa = 9.45NNVTIKK206 pKa = 9.41TQTNANQIYY215 pKa = 8.12WWFNPVRR222 pKa = 11.84DD223 pKa = 3.67LSYY226 pKa = 10.96YY227 pKa = 10.35NVNKK231 pKa = 9.63TVDD234 pKa = 3.4STIVVSNCTSDD245 pKa = 3.43CSGYY249 pKa = 10.39AANIFAVEE257 pKa = 4.12TGGFIPSSFSFNNWFVLTNSSTIVSGKK284 pKa = 9.78FVSSQPLLVNCLTPVPSFGDD304 pKa = 3.31EE305 pKa = 3.8TSIINFDD312 pKa = 4.06TVPSQCNGATVNGSFDD328 pKa = 3.49VVRR331 pKa = 11.84FNLNFTANVASSSGTSFLALNTTGGVVLYY360 pKa = 8.29LSCFNEE366 pKa = 4.25TKK368 pKa = 8.19TTNLYY373 pKa = 9.43MSEE376 pKa = 4.06GALPFGTHH384 pKa = 6.29EE385 pKa = 4.34GALYY389 pKa = 10.53CYY391 pKa = 10.38VSYY394 pKa = 11.56NEE396 pKa = 4.29TSFKK400 pKa = 10.75YY401 pKa = 10.14IGVLPPSVKK410 pKa = 10.29EE411 pKa = 3.65IAVSKK416 pKa = 10.09WGFVYY421 pKa = 10.59INGYY425 pKa = 10.63NYY427 pKa = 10.16FQTFPIDD434 pKa = 3.43SVAFNLTTGNSGAFWTIAYY453 pKa = 7.01TTFTDD458 pKa = 3.4VLLEE462 pKa = 4.21VSDD465 pKa = 3.99TQIKK469 pKa = 10.22SVTYY473 pKa = 10.18CNSHH477 pKa = 6.99INDD480 pKa = 3.81IKK482 pKa = 10.42CSQMSEE488 pKa = 4.0NLPDD492 pKa = 3.23GFYY495 pKa = 10.52PVSQHH500 pKa = 6.53SLPNVNKK507 pKa = 9.6TFVTLPANFEE517 pKa = 4.14HH518 pKa = 6.76TSINVTGNVLLIYY531 pKa = 9.48YY532 pKa = 7.1AHH534 pKa = 6.84PVFRR538 pKa = 11.84SGNVTLHH545 pKa = 5.83PQGTQTICVNTTQFTFNFDD564 pKa = 4.0SQCHH568 pKa = 4.18QLAGGGCDD576 pKa = 3.81GIGASMISIDD586 pKa = 4.92SGNCPFSPDD595 pKa = 4.12KK596 pKa = 11.18LNQHH600 pKa = 6.75LAFEE604 pKa = 4.49TLCFSTLPTGSDD616 pKa = 3.54CAFSLTVQNRR626 pKa = 11.84YY627 pKa = 5.17FTRR630 pKa = 11.84VFAHH634 pKa = 6.68LFVTYY639 pKa = 10.22KK640 pKa = 10.79YY641 pKa = 10.67GLDD644 pKa = 3.38HH645 pKa = 7.22LGVQTPDD652 pKa = 3.11VGVKK656 pKa = 10.15DD657 pKa = 4.02LSVVYY662 pKa = 9.85QNVCTEE668 pKa = 3.84YY669 pKa = 10.84NIYY672 pKa = 10.17GHH674 pKa = 7.04AGVGIIRR681 pKa = 11.84STNQTLLGGLYY692 pKa = 8.33YY693 pKa = 10.54TSLSGDD699 pKa = 3.35LLGFKK704 pKa = 10.35NVTTGEE710 pKa = 4.27VYY712 pKa = 10.74SIVPCQLSAQAAVINGKK729 pKa = 9.35IVGAVTSVQSPILDD743 pKa = 3.96LPHH746 pKa = 7.16HH747 pKa = 6.41IVTPQFYY754 pKa = 9.64YY755 pKa = 10.62HH756 pKa = 7.09SIYY759 pKa = 10.65NYY761 pKa = 9.87SATPTSYY768 pKa = 9.3RR769 pKa = 11.84TNNGFDD775 pKa = 3.47KK776 pKa = 11.17YY777 pKa = 10.98LVNCTPIITYY787 pKa = 10.93SNMGVCEE794 pKa = 3.87NGALVFINITQSEE807 pKa = 4.64NPVQPISTGNVTIPSNFTISVQVEE831 pKa = 3.96YY832 pKa = 11.03LQMSSEE838 pKa = 4.25AVSIDD843 pKa = 3.18CAQYY847 pKa = 10.24VCNGNPRR854 pKa = 11.84CNRR857 pKa = 11.84LLAQYY862 pKa = 10.42ISACHH867 pKa = 6.55AIEE870 pKa = 3.9QALITSTRR878 pKa = 11.84LEE880 pKa = 4.05ALEE883 pKa = 4.53LEE885 pKa = 4.52SMISISDD892 pKa = 3.57NALALATVEE901 pKa = 4.52TFNSSDD907 pKa = 3.63YY908 pKa = 11.15LDD910 pKa = 3.95PVYY913 pKa = 10.96NDD915 pKa = 3.1QHH917 pKa = 5.3NTIGGIYY924 pKa = 9.08MDD926 pKa = 4.39GLKK929 pKa = 10.72DD930 pKa = 3.47LLPRR934 pKa = 11.84RR935 pKa = 11.84SCANKK940 pKa = 9.99HH941 pKa = 4.29GTCRR945 pKa = 11.84SVIEE949 pKa = 3.89EE950 pKa = 4.11LLFNKK955 pKa = 9.48VVTSGLGTVDD965 pKa = 3.79EE966 pKa = 4.9DD967 pKa = 4.87YY968 pKa = 11.24KK969 pKa = 11.15RR970 pKa = 11.84CTNGLDD976 pKa = 3.62IADD979 pKa = 4.62LVCAQYY985 pKa = 11.58YY986 pKa = 8.99NGIMVLPGVVNADD999 pKa = 3.0KK1000 pKa = 10.45MAMYY1004 pKa = 8.05TASLAGGITLGALGGGLVSVPFATAVQARR1033 pKa = 11.84LNYY1036 pKa = 9.72VALQTDD1042 pKa = 4.22VLQQNQKK1049 pKa = 9.71ILAASFNQAIGNITLAFGKK1068 pKa = 10.32VNSAIQQTAQGLSTVAQALTKK1089 pKa = 10.33VQDD1092 pKa = 3.72VVNSQGKK1099 pKa = 9.12ALNHH1103 pKa = 5.94LTAQLQNNFQAISSSIEE1120 pKa = 3.99DD1121 pKa = 3.12IYY1123 pKa = 11.76YY1124 pKa = 10.95KK1125 pKa = 10.47LDD1127 pKa = 3.56EE1128 pKa = 4.62VNADD1132 pKa = 3.46AQVDD1136 pKa = 3.81RR1137 pKa = 11.84LITGRR1142 pKa = 11.84LASLNAFVTQTLTNQAQIRR1161 pKa = 11.84ASRR1164 pKa = 11.84QLSKK1168 pKa = 11.24EE1169 pKa = 4.03KK1170 pKa = 10.5INEE1173 pKa = 4.27CVRR1176 pKa = 11.84SQSSRR1181 pKa = 11.84FGFCGNGTHH1190 pKa = 7.49LFSLANAAPRR1200 pKa = 11.84GVMLFHH1206 pKa = 6.51TVLLPTSYY1214 pKa = 9.07KK1215 pKa = 10.18TVTAWSGVCAISNNKK1230 pKa = 8.2TMGFIVKK1237 pKa = 9.91DD1238 pKa = 3.55VSLTLFKK1245 pKa = 11.26NHH1247 pKa = 7.23DD1248 pKa = 3.99DD1249 pKa = 3.9KK1250 pKa = 11.61FYY1252 pKa = 9.47LTPRR1256 pKa = 11.84TMYY1259 pKa = 9.32EE1260 pKa = 3.65PRR1262 pKa = 11.84VATMSDD1268 pKa = 3.53FVQIEE1273 pKa = 4.54SCTTTFVNATVAEE1286 pKa = 4.51LPSIIPDD1293 pKa = 3.78YY1294 pKa = 11.1IDD1296 pKa = 3.31INGTIKK1302 pKa = 11.04DD1303 pKa = 3.94MLDD1306 pKa = 3.04QYY1308 pKa = 10.89KK1309 pKa = 10.44PNWTVPEE1316 pKa = 3.94LTIDD1320 pKa = 3.94VFNQTYY1326 pKa = 10.97LNLTGEE1332 pKa = 4.32INEE1335 pKa = 4.23LEE1337 pKa = 4.13NRR1339 pKa = 11.84SVILQQTVLEE1349 pKa = 4.5LEE1351 pKa = 4.27SLIDD1355 pKa = 4.28NINGTLVNLEE1365 pKa = 3.71WLNRR1369 pKa = 11.84VEE1371 pKa = 5.64TFVKK1375 pKa = 8.87WPWWVWVIIGLILLIALPMLLFCCLSTGCCGCCGCLTSCLAGCCKK1420 pKa = 10.33NSCKK1424 pKa = 10.19RR1425 pKa = 11.84PSYY1428 pKa = 9.43YY1429 pKa = 10.55EE1430 pKa = 3.75PMEE1433 pKa = 4.32KK1434 pKa = 10.25VHH1436 pKa = 7.08INN1438 pKa = 3.15
Molecular weight: 157.98 kDa
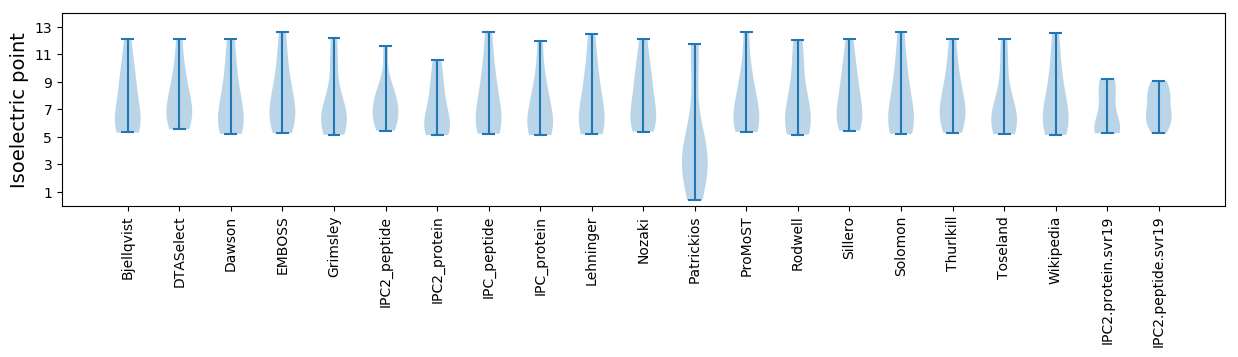
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A172B5K4|A0A172B5K4_9ALPC Envelope small membrane protein OS=Ferret coronavirus OX=1264898 GN=E PE=3 SV=1
MM1 pKa = 7.48SSSLITIFSGKK12 pKa = 8.72IWFSLSRR19 pKa = 11.84PFKK22 pKa = 10.06DD23 pKa = 2.98WIVSKK28 pKa = 11.02VRR30 pKa = 11.84FKK32 pKa = 10.96TPVGGKK38 pKa = 9.67VKK40 pKa = 10.48LDD42 pKa = 3.27YY43 pKa = 10.5RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84ALLNSHH52 pKa = 6.23NNHH55 pKa = 5.52VNSNFNSSVSVLKK68 pKa = 9.88FLRR71 pKa = 11.84ARR73 pKa = 11.84RR74 pKa = 11.84WQASTSHH81 pKa = 5.81IQLGKK86 pKa = 10.2IRR88 pKa = 11.84VFF90 pKa = 3.62
MM1 pKa = 7.48SSSLITIFSGKK12 pKa = 8.72IWFSLSRR19 pKa = 11.84PFKK22 pKa = 10.06DD23 pKa = 2.98WIVSKK28 pKa = 11.02VRR30 pKa = 11.84FKK32 pKa = 10.96TPVGGKK38 pKa = 9.67VKK40 pKa = 10.48LDD42 pKa = 3.27YY43 pKa = 10.5RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84ALLNSHH52 pKa = 6.23NNHH55 pKa = 5.52VNSNFNSSVSVLKK68 pKa = 9.88FLRR71 pKa = 11.84ARR73 pKa = 11.84RR74 pKa = 11.84WQASTSHH81 pKa = 5.81IQLGKK86 pKa = 10.2IRR88 pKa = 11.84VFF90 pKa = 3.62
Molecular weight: 10.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
13279 |
82 |
6622 |
1475.4 |
164.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.461 ± 0.26 | 3.276 ± 0.357 |
5.543 ± 0.461 | 3.961 ± 0.211 |
5.452 ± 0.264 | 6.657 ± 0.233 |
1.86 ± 0.078 | 4.654 ± 0.46 |
5.859 ± 0.677 | 8.962 ± 0.321 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.47 ± 0.327 | 5.889 ± 0.452 |
3.457 ± 0.191 | 2.734 ± 0.372 |
3.268 ± 0.493 | 7.214 ± 0.541 |
5.874 ± 0.62 | 10.264 ± 0.657 |
1.205 ± 0.226 | 4.94 ± 0.276 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
