
Hubei virga-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
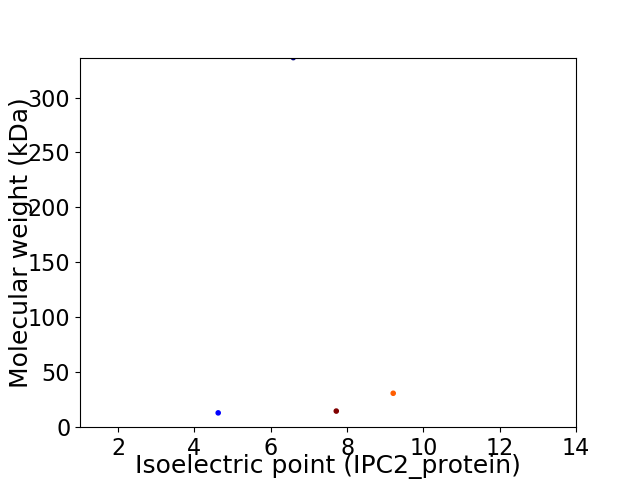
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KK45|A0A1L3KK45_9VIRU Uncharacterized protein OS=Hubei virga-like virus 9 OX=1923342 PE=4 SV=1
MM1 pKa = 7.64CEE3 pKa = 4.39KK4 pKa = 10.24SDD6 pKa = 3.48SGIIGEE12 pKa = 4.09EE13 pKa = 4.1SYY15 pKa = 11.21GIEE18 pKa = 3.37RR19 pKa = 11.84LLEE22 pKa = 4.0NEE24 pKa = 4.68FIHH27 pKa = 7.32YY28 pKa = 10.75DD29 pKa = 4.09LDD31 pKa = 4.79DD32 pKa = 4.3KK33 pKa = 11.26LQQRR37 pKa = 11.84LVITLLLTNSWRR49 pKa = 11.84SDD51 pKa = 4.34PIWLLQKK58 pKa = 9.16ITAARR63 pKa = 11.84KK64 pKa = 7.93RR65 pKa = 11.84DD66 pKa = 3.39TYY68 pKa = 11.51SFYY71 pKa = 10.3EE72 pKa = 4.18SRR74 pKa = 11.84WFEE77 pKa = 3.97IEE79 pKa = 4.88AGGQQATTDD88 pKa = 3.6GSYY91 pKa = 11.08QDD93 pKa = 3.49NTEE96 pKa = 3.76EE97 pKa = 4.98GEE99 pKa = 4.15THH101 pKa = 6.33KK102 pKa = 10.63RR103 pKa = 11.84GRR105 pKa = 11.84YY106 pKa = 9.41DD107 pKa = 4.84DD108 pKa = 4.17YY109 pKa = 11.88SCC111 pKa = 5.25
MM1 pKa = 7.64CEE3 pKa = 4.39KK4 pKa = 10.24SDD6 pKa = 3.48SGIIGEE12 pKa = 4.09EE13 pKa = 4.1SYY15 pKa = 11.21GIEE18 pKa = 3.37RR19 pKa = 11.84LLEE22 pKa = 4.0NEE24 pKa = 4.68FIHH27 pKa = 7.32YY28 pKa = 10.75DD29 pKa = 4.09LDD31 pKa = 4.79DD32 pKa = 4.3KK33 pKa = 11.26LQQRR37 pKa = 11.84LVITLLLTNSWRR49 pKa = 11.84SDD51 pKa = 4.34PIWLLQKK58 pKa = 9.16ITAARR63 pKa = 11.84KK64 pKa = 7.93RR65 pKa = 11.84DD66 pKa = 3.39TYY68 pKa = 11.51SFYY71 pKa = 10.3EE72 pKa = 4.18SRR74 pKa = 11.84WFEE77 pKa = 3.97IEE79 pKa = 4.88AGGQQATTDD88 pKa = 3.6GSYY91 pKa = 11.08QDD93 pKa = 3.49NTEE96 pKa = 3.76EE97 pKa = 4.98GEE99 pKa = 4.15THH101 pKa = 6.33KK102 pKa = 10.63RR103 pKa = 11.84GRR105 pKa = 11.84YY106 pKa = 9.41DD107 pKa = 4.84DD108 pKa = 4.17YY109 pKa = 11.88SCC111 pKa = 5.25
Molecular weight: 13.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJW2|A0A1L3KJW2_9VIRU Uncharacterized protein OS=Hubei virga-like virus 9 OX=1923342 PE=4 SV=1
MM1 pKa = 7.21SHH3 pKa = 6.39GGSRR7 pKa = 11.84SRR9 pKa = 11.84QAANKK14 pKa = 9.42RR15 pKa = 11.84RR16 pKa = 11.84QTGHH20 pKa = 6.22TKK22 pKa = 10.05TIPKK26 pKa = 9.82RR27 pKa = 11.84VRR29 pKa = 11.84HH30 pKa = 5.07TSEE33 pKa = 3.93GDD35 pKa = 3.22TTIILADD42 pKa = 3.85SPEE45 pKa = 4.39GTSTRR50 pKa = 11.84TPSKK54 pKa = 9.95RR55 pKa = 11.84ARR57 pKa = 11.84HH58 pKa = 5.24VSASATITTPIEE70 pKa = 4.0TPEE73 pKa = 4.13EE74 pKa = 4.51YY75 pKa = 9.01NTSNLTVDD83 pKa = 4.68PSPSSPEE90 pKa = 3.81PQFTPPSAIVSRR102 pKa = 11.84SVRR105 pKa = 11.84IRR107 pKa = 11.84GRR109 pKa = 11.84SLDD112 pKa = 3.57MTTPKK117 pKa = 10.48YY118 pKa = 10.31EE119 pKa = 3.74RR120 pKa = 11.84WTSRR124 pKa = 11.84MEE126 pKa = 4.14RR127 pKa = 11.84YY128 pKa = 8.38TNANFMDD135 pKa = 3.88AVIVEE140 pKa = 4.23DD141 pKa = 3.91TLRR144 pKa = 11.84EE145 pKa = 3.92MSVRR149 pKa = 11.84NYY151 pKa = 10.28SNRR154 pKa = 11.84PARR157 pKa = 11.84EE158 pKa = 3.81SALTSIVSMNGDD170 pKa = 3.91RR171 pKa = 11.84IVGPTLRR178 pKa = 11.84FPDD181 pKa = 3.44TGVYY185 pKa = 9.99ILVSTGPLRR194 pKa = 11.84EE195 pKa = 4.57LFDD198 pKa = 4.59QIILACTLTEE208 pKa = 4.21SDD210 pKa = 4.05PTTSSGSAHH219 pKa = 6.25NRR221 pKa = 11.84YY222 pKa = 9.97DD223 pKa = 3.89DD224 pKa = 4.34NKK226 pKa = 10.3LSYY229 pKa = 10.92YY230 pKa = 10.32KK231 pKa = 10.53AISNALFLLRR241 pKa = 11.84RR242 pKa = 11.84RR243 pKa = 11.84FEE245 pKa = 4.38PAAHH249 pKa = 6.38VYY251 pKa = 10.36DD252 pKa = 3.68IYY254 pKa = 11.45SRR256 pKa = 11.84STFEE260 pKa = 3.74STNRR264 pKa = 11.84IVWNDD269 pKa = 3.0GNTTRR274 pKa = 11.84QQ275 pKa = 3.13
MM1 pKa = 7.21SHH3 pKa = 6.39GGSRR7 pKa = 11.84SRR9 pKa = 11.84QAANKK14 pKa = 9.42RR15 pKa = 11.84RR16 pKa = 11.84QTGHH20 pKa = 6.22TKK22 pKa = 10.05TIPKK26 pKa = 9.82RR27 pKa = 11.84VRR29 pKa = 11.84HH30 pKa = 5.07TSEE33 pKa = 3.93GDD35 pKa = 3.22TTIILADD42 pKa = 3.85SPEE45 pKa = 4.39GTSTRR50 pKa = 11.84TPSKK54 pKa = 9.95RR55 pKa = 11.84ARR57 pKa = 11.84HH58 pKa = 5.24VSASATITTPIEE70 pKa = 4.0TPEE73 pKa = 4.13EE74 pKa = 4.51YY75 pKa = 9.01NTSNLTVDD83 pKa = 4.68PSPSSPEE90 pKa = 3.81PQFTPPSAIVSRR102 pKa = 11.84SVRR105 pKa = 11.84IRR107 pKa = 11.84GRR109 pKa = 11.84SLDD112 pKa = 3.57MTTPKK117 pKa = 10.48YY118 pKa = 10.31EE119 pKa = 3.74RR120 pKa = 11.84WTSRR124 pKa = 11.84MEE126 pKa = 4.14RR127 pKa = 11.84YY128 pKa = 8.38TNANFMDD135 pKa = 3.88AVIVEE140 pKa = 4.23DD141 pKa = 3.91TLRR144 pKa = 11.84EE145 pKa = 3.92MSVRR149 pKa = 11.84NYY151 pKa = 10.28SNRR154 pKa = 11.84PARR157 pKa = 11.84EE158 pKa = 3.81SALTSIVSMNGDD170 pKa = 3.91RR171 pKa = 11.84IVGPTLRR178 pKa = 11.84FPDD181 pKa = 3.44TGVYY185 pKa = 9.99ILVSTGPLRR194 pKa = 11.84EE195 pKa = 4.57LFDD198 pKa = 4.59QIILACTLTEE208 pKa = 4.21SDD210 pKa = 4.05PTTSSGSAHH219 pKa = 6.25NRR221 pKa = 11.84YY222 pKa = 9.97DD223 pKa = 3.89DD224 pKa = 4.34NKK226 pKa = 10.3LSYY229 pKa = 10.92YY230 pKa = 10.32KK231 pKa = 10.53AISNALFLLRR241 pKa = 11.84RR242 pKa = 11.84RR243 pKa = 11.84FEE245 pKa = 4.38PAAHH249 pKa = 6.38VYY251 pKa = 10.36DD252 pKa = 3.68IYY254 pKa = 11.45SRR256 pKa = 11.84STFEE260 pKa = 3.74STNRR264 pKa = 11.84IVWNDD269 pKa = 3.0GNTTRR274 pKa = 11.84QQ275 pKa = 3.13
Molecular weight: 30.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3460 |
111 |
2947 |
865.0 |
98.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.04 ± 1.061 | 2.312 ± 0.79 |
7.254 ± 0.659 | 5.405 ± 0.972 |
3.757 ± 0.697 | 4.798 ± 0.441 |
2.688 ± 0.336 | 5.145 ± 0.434 |
5.867 ± 0.944 | 8.41 ± 0.872 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.642 ± 0.602 | 4.191 ± 0.376 |
4.104 ± 1.348 | 2.312 ± 0.57 |
6.821 ± 0.93 | 7.081 ± 1.225 |
6.908 ± 1.482 | 7.63 ± 2.14 |
1.04 ± 0.3 | 4.595 ± 0.484 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
