
Cocksfoot mottle virus (isolate Dactylis glomerata/Norway/CfMV-NO/1995) (CfMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus; Cocksfoot mottle virus
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
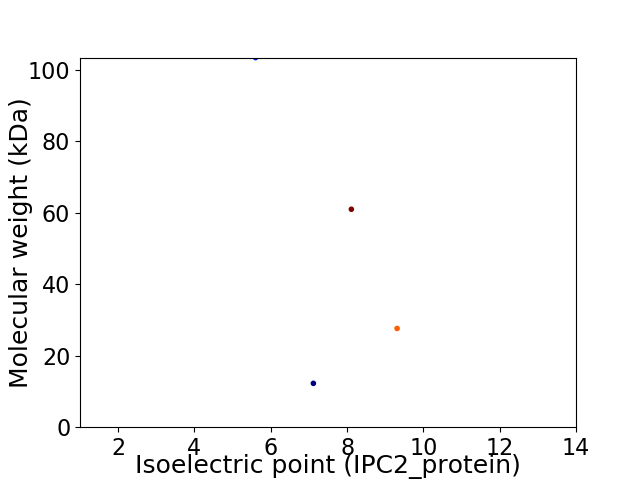
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q66011|MVP_CFMVN Movement protein P1 OS=Cocksfoot mottle virus (isolate Dactylis glomerata/Norway/CfMV-NO/1995) OX=1005059 GN=ORF1 PE=4 SV=1
MM1 pKa = 7.12GCSVVGNCKK10 pKa = 10.22SVMLMSRR17 pKa = 11.84MSWSKK22 pKa = 10.4LALLISVAMAAAMTDD37 pKa = 3.45SPPTLICMGILVSVVLNWIVCAVCEE62 pKa = 4.13EE63 pKa = 4.03ASEE66 pKa = 4.89LILGVSLEE74 pKa = 4.33TTRR77 pKa = 11.84PSPARR82 pKa = 11.84VIGEE86 pKa = 4.05PVFDD90 pKa = 3.82PRR92 pKa = 11.84YY93 pKa = 9.97GYY95 pKa = 9.65VAPAIYY101 pKa = 9.85DD102 pKa = 3.42GKK104 pKa = 10.85SFDD107 pKa = 4.03VILPISALSSASTRR121 pKa = 11.84KK122 pKa = 8.39EE123 pKa = 4.16TVEE126 pKa = 3.8MAVEE130 pKa = 4.16NSRR133 pKa = 11.84LQPLEE138 pKa = 3.92SSQTPKK144 pKa = 11.0SLVALYY150 pKa = 10.59SQDD153 pKa = 4.54LLSGWGSRR161 pKa = 11.84IKK163 pKa = 10.99GPDD166 pKa = 3.24GQEE169 pKa = 3.87YY170 pKa = 10.72LLTALHH176 pKa = 5.71VWEE179 pKa = 4.79TNISHH184 pKa = 7.22LCKK187 pKa = 10.25DD188 pKa = 3.73GKK190 pKa = 10.3KK191 pKa = 10.14VPISGCPIVASSADD205 pKa = 3.39SDD207 pKa = 4.12LDD209 pKa = 3.75FVLVSVPKK217 pKa = 10.35NAWSVLGVGVARR229 pKa = 11.84LEE231 pKa = 3.93LLKK234 pKa = 10.63RR235 pKa = 11.84RR236 pKa = 11.84TVVTVYY242 pKa = 10.92GGLDD246 pKa = 3.2SKK248 pKa = 8.32TTYY251 pKa = 10.25CATGVAEE258 pKa = 4.57LEE260 pKa = 4.04NPFRR264 pKa = 11.84IVTKK268 pKa = 9.92VTTTGGWSGSPLYY281 pKa = 10.76HH282 pKa = 7.08KK283 pKa = 10.5DD284 pKa = 3.45AIVGLHH290 pKa = 6.22LGARR294 pKa = 11.84PSAGVNRR301 pKa = 11.84ACNVAMAFRR310 pKa = 11.84VVRR313 pKa = 11.84KK314 pKa = 9.01FVTVEE319 pKa = 3.65NSEE322 pKa = 4.7LYY324 pKa = 9.86PDD326 pKa = 3.99QSSGPARR333 pKa = 11.84EE334 pKa = 4.59LDD336 pKa = 3.18AEE338 pKa = 4.74TYY340 pKa = 8.52TEE342 pKa = 4.23RR343 pKa = 11.84LEE345 pKa = 3.88QGIAFTEE352 pKa = 4.32YY353 pKa = 9.71NISGITVKK361 pKa = 10.24TSDD364 pKa = 4.4RR365 pKa = 11.84EE366 pKa = 4.31WTTAEE371 pKa = 3.86ALRR374 pKa = 11.84VARR377 pKa = 11.84YY378 pKa = 9.41KK379 pKa = 10.78PLGGGKK385 pKa = 9.65AWGDD389 pKa = 3.51SDD391 pKa = 6.47DD392 pKa = 5.18EE393 pKa = 4.48DD394 pKa = 4.09TQEE397 pKa = 4.01TAIRR401 pKa = 11.84PLNLPAGGLPTGQSALGQLIEE422 pKa = 4.08YY423 pKa = 9.95AGYY426 pKa = 8.61VWRR429 pKa = 11.84DD430 pKa = 3.16EE431 pKa = 5.22GIINSNGMPFRR442 pKa = 11.84SAGKK446 pKa = 8.13SSCRR450 pKa = 11.84FRR452 pKa = 11.84EE453 pKa = 4.21AVCRR457 pKa = 11.84AVHH460 pKa = 6.16RR461 pKa = 11.84DD462 pKa = 2.91VRR464 pKa = 11.84AAEE467 pKa = 4.23TEE469 pKa = 4.05FPEE472 pKa = 4.77LKK474 pKa = 10.23EE475 pKa = 3.84LAWPSRR481 pKa = 11.84GSKK484 pKa = 10.85AEE486 pKa = 3.79IGSLLFQAGRR496 pKa = 11.84FEE498 pKa = 4.83RR499 pKa = 11.84VEE501 pKa = 4.14APANLQLAITNLQAQYY517 pKa = 10.05PRR519 pKa = 11.84SRR521 pKa = 11.84PRR523 pKa = 11.84SCFRR527 pKa = 11.84RR528 pKa = 11.84EE529 pKa = 3.51PWCRR533 pKa = 11.84EE534 pKa = 3.76DD535 pKa = 4.88FVAEE539 pKa = 4.0IEE541 pKa = 4.64KK542 pKa = 10.04IAHH545 pKa = 5.75SGEE548 pKa = 4.31INLKK552 pKa = 10.26ASPGVPLAEE561 pKa = 4.11IGVSNQQVIDD571 pKa = 3.87VAWPLVCEE579 pKa = 4.11AVVEE583 pKa = 4.22RR584 pKa = 11.84LHH586 pKa = 7.3ALASVDD592 pKa = 3.71PRR594 pKa = 11.84QHH596 pKa = 6.25DD597 pKa = 3.55WSPEE601 pKa = 3.83EE602 pKa = 3.72LVKK605 pKa = 10.77RR606 pKa = 11.84GLCDD610 pKa = 3.38PVRR613 pKa = 11.84LFVKK617 pKa = 10.06QEE619 pKa = 3.43PHH621 pKa = 5.65SRR623 pKa = 11.84QKK625 pKa = 10.34IEE627 pKa = 3.55QGRR630 pKa = 11.84FRR632 pKa = 11.84LISSVSLVDD641 pKa = 3.53QLVEE645 pKa = 4.17RR646 pKa = 11.84MLFGPQNTTEE656 pKa = 3.72IALWHH661 pKa = 6.4SNPSKK666 pKa = 10.65PGMGLSKK673 pKa = 10.66ASQVALLWEE682 pKa = 4.51DD683 pKa = 4.16LARR686 pKa = 11.84KK687 pKa = 8.3HH688 pKa = 4.78QTHH691 pKa = 7.31PGAMADD697 pKa = 2.91ISGFDD702 pKa = 3.38WSVQDD707 pKa = 3.34WEE709 pKa = 4.3LWADD713 pKa = 3.04VSMRR717 pKa = 11.84IEE719 pKa = 4.55LGSFPALMAKK729 pKa = 10.19AAISRR734 pKa = 11.84FYY736 pKa = 11.27CLMNATFQLTNGEE749 pKa = 4.81LLTQEE754 pKa = 5.21LPGLMKK760 pKa = 10.55SGSYY764 pKa = 8.58CTSSSNSRR772 pKa = 11.84IRR774 pKa = 11.84CLMAEE779 pKa = 4.85LIGSPWCIAMGDD791 pKa = 3.8DD792 pKa = 5.11SVEE795 pKa = 3.99GWVDD799 pKa = 3.48DD800 pKa = 4.52APRR803 pKa = 11.84KK804 pKa = 9.78YY805 pKa = 10.42SALGHH810 pKa = 6.05LCKK813 pKa = 10.17EE814 pKa = 4.42YY815 pKa = 10.26EE816 pKa = 4.36ACPVLPNGDD825 pKa = 3.52LKK827 pKa = 11.05EE828 pKa = 4.18VSFCSHH834 pKa = 7.62LISKK838 pKa = 9.66GRR840 pKa = 11.84AEE842 pKa = 4.34LEE844 pKa = 4.02TWPKK848 pKa = 10.65CLFRR852 pKa = 11.84YY853 pKa = 10.24LSGPHH858 pKa = 6.57DD859 pKa = 4.0VEE861 pKa = 4.25SLEE864 pKa = 4.39MEE866 pKa = 4.61LSSSRR871 pKa = 11.84RR872 pKa = 11.84WGQIVRR878 pKa = 11.84YY879 pKa = 8.48LRR881 pKa = 11.84RR882 pKa = 11.84IGRR885 pKa = 11.84VSGNDD890 pKa = 3.46GEE892 pKa = 4.54EE893 pKa = 3.97RR894 pKa = 11.84SSNEE898 pKa = 3.59SPATTKK904 pKa = 9.49TQGSAAAWGPPQEE917 pKa = 4.16AWPVDD922 pKa = 3.66GASLSTFEE930 pKa = 4.94PSSSGWFHH938 pKa = 7.76LEE940 pKa = 3.5GWW942 pKa = 3.69
MM1 pKa = 7.12GCSVVGNCKK10 pKa = 10.22SVMLMSRR17 pKa = 11.84MSWSKK22 pKa = 10.4LALLISVAMAAAMTDD37 pKa = 3.45SPPTLICMGILVSVVLNWIVCAVCEE62 pKa = 4.13EE63 pKa = 4.03ASEE66 pKa = 4.89LILGVSLEE74 pKa = 4.33TTRR77 pKa = 11.84PSPARR82 pKa = 11.84VIGEE86 pKa = 4.05PVFDD90 pKa = 3.82PRR92 pKa = 11.84YY93 pKa = 9.97GYY95 pKa = 9.65VAPAIYY101 pKa = 9.85DD102 pKa = 3.42GKK104 pKa = 10.85SFDD107 pKa = 4.03VILPISALSSASTRR121 pKa = 11.84KK122 pKa = 8.39EE123 pKa = 4.16TVEE126 pKa = 3.8MAVEE130 pKa = 4.16NSRR133 pKa = 11.84LQPLEE138 pKa = 3.92SSQTPKK144 pKa = 11.0SLVALYY150 pKa = 10.59SQDD153 pKa = 4.54LLSGWGSRR161 pKa = 11.84IKK163 pKa = 10.99GPDD166 pKa = 3.24GQEE169 pKa = 3.87YY170 pKa = 10.72LLTALHH176 pKa = 5.71VWEE179 pKa = 4.79TNISHH184 pKa = 7.22LCKK187 pKa = 10.25DD188 pKa = 3.73GKK190 pKa = 10.3KK191 pKa = 10.14VPISGCPIVASSADD205 pKa = 3.39SDD207 pKa = 4.12LDD209 pKa = 3.75FVLVSVPKK217 pKa = 10.35NAWSVLGVGVARR229 pKa = 11.84LEE231 pKa = 3.93LLKK234 pKa = 10.63RR235 pKa = 11.84RR236 pKa = 11.84TVVTVYY242 pKa = 10.92GGLDD246 pKa = 3.2SKK248 pKa = 8.32TTYY251 pKa = 10.25CATGVAEE258 pKa = 4.57LEE260 pKa = 4.04NPFRR264 pKa = 11.84IVTKK268 pKa = 9.92VTTTGGWSGSPLYY281 pKa = 10.76HH282 pKa = 7.08KK283 pKa = 10.5DD284 pKa = 3.45AIVGLHH290 pKa = 6.22LGARR294 pKa = 11.84PSAGVNRR301 pKa = 11.84ACNVAMAFRR310 pKa = 11.84VVRR313 pKa = 11.84KK314 pKa = 9.01FVTVEE319 pKa = 3.65NSEE322 pKa = 4.7LYY324 pKa = 9.86PDD326 pKa = 3.99QSSGPARR333 pKa = 11.84EE334 pKa = 4.59LDD336 pKa = 3.18AEE338 pKa = 4.74TYY340 pKa = 8.52TEE342 pKa = 4.23RR343 pKa = 11.84LEE345 pKa = 3.88QGIAFTEE352 pKa = 4.32YY353 pKa = 9.71NISGITVKK361 pKa = 10.24TSDD364 pKa = 4.4RR365 pKa = 11.84EE366 pKa = 4.31WTTAEE371 pKa = 3.86ALRR374 pKa = 11.84VARR377 pKa = 11.84YY378 pKa = 9.41KK379 pKa = 10.78PLGGGKK385 pKa = 9.65AWGDD389 pKa = 3.51SDD391 pKa = 6.47DD392 pKa = 5.18EE393 pKa = 4.48DD394 pKa = 4.09TQEE397 pKa = 4.01TAIRR401 pKa = 11.84PLNLPAGGLPTGQSALGQLIEE422 pKa = 4.08YY423 pKa = 9.95AGYY426 pKa = 8.61VWRR429 pKa = 11.84DD430 pKa = 3.16EE431 pKa = 5.22GIINSNGMPFRR442 pKa = 11.84SAGKK446 pKa = 8.13SSCRR450 pKa = 11.84FRR452 pKa = 11.84EE453 pKa = 4.21AVCRR457 pKa = 11.84AVHH460 pKa = 6.16RR461 pKa = 11.84DD462 pKa = 2.91VRR464 pKa = 11.84AAEE467 pKa = 4.23TEE469 pKa = 4.05FPEE472 pKa = 4.77LKK474 pKa = 10.23EE475 pKa = 3.84LAWPSRR481 pKa = 11.84GSKK484 pKa = 10.85AEE486 pKa = 3.79IGSLLFQAGRR496 pKa = 11.84FEE498 pKa = 4.83RR499 pKa = 11.84VEE501 pKa = 4.14APANLQLAITNLQAQYY517 pKa = 10.05PRR519 pKa = 11.84SRR521 pKa = 11.84PRR523 pKa = 11.84SCFRR527 pKa = 11.84RR528 pKa = 11.84EE529 pKa = 3.51PWCRR533 pKa = 11.84EE534 pKa = 3.76DD535 pKa = 4.88FVAEE539 pKa = 4.0IEE541 pKa = 4.64KK542 pKa = 10.04IAHH545 pKa = 5.75SGEE548 pKa = 4.31INLKK552 pKa = 10.26ASPGVPLAEE561 pKa = 4.11IGVSNQQVIDD571 pKa = 3.87VAWPLVCEE579 pKa = 4.11AVVEE583 pKa = 4.22RR584 pKa = 11.84LHH586 pKa = 7.3ALASVDD592 pKa = 3.71PRR594 pKa = 11.84QHH596 pKa = 6.25DD597 pKa = 3.55WSPEE601 pKa = 3.83EE602 pKa = 3.72LVKK605 pKa = 10.77RR606 pKa = 11.84GLCDD610 pKa = 3.38PVRR613 pKa = 11.84LFVKK617 pKa = 10.06QEE619 pKa = 3.43PHH621 pKa = 5.65SRR623 pKa = 11.84QKK625 pKa = 10.34IEE627 pKa = 3.55QGRR630 pKa = 11.84FRR632 pKa = 11.84LISSVSLVDD641 pKa = 3.53QLVEE645 pKa = 4.17RR646 pKa = 11.84MLFGPQNTTEE656 pKa = 3.72IALWHH661 pKa = 6.4SNPSKK666 pKa = 10.65PGMGLSKK673 pKa = 10.66ASQVALLWEE682 pKa = 4.51DD683 pKa = 4.16LARR686 pKa = 11.84KK687 pKa = 8.3HH688 pKa = 4.78QTHH691 pKa = 7.31PGAMADD697 pKa = 2.91ISGFDD702 pKa = 3.38WSVQDD707 pKa = 3.34WEE709 pKa = 4.3LWADD713 pKa = 3.04VSMRR717 pKa = 11.84IEE719 pKa = 4.55LGSFPALMAKK729 pKa = 10.19AAISRR734 pKa = 11.84FYY736 pKa = 11.27CLMNATFQLTNGEE749 pKa = 4.81LLTQEE754 pKa = 5.21LPGLMKK760 pKa = 10.55SGSYY764 pKa = 8.58CTSSSNSRR772 pKa = 11.84IRR774 pKa = 11.84CLMAEE779 pKa = 4.85LIGSPWCIAMGDD791 pKa = 3.8DD792 pKa = 5.11SVEE795 pKa = 3.99GWVDD799 pKa = 3.48DD800 pKa = 4.52APRR803 pKa = 11.84KK804 pKa = 9.78YY805 pKa = 10.42SALGHH810 pKa = 6.05LCKK813 pKa = 10.17EE814 pKa = 4.42YY815 pKa = 10.26EE816 pKa = 4.36ACPVLPNGDD825 pKa = 3.52LKK827 pKa = 11.05EE828 pKa = 4.18VSFCSHH834 pKa = 7.62LISKK838 pKa = 9.66GRR840 pKa = 11.84AEE842 pKa = 4.34LEE844 pKa = 4.02TWPKK848 pKa = 10.65CLFRR852 pKa = 11.84YY853 pKa = 10.24LSGPHH858 pKa = 6.57DD859 pKa = 4.0VEE861 pKa = 4.25SLEE864 pKa = 4.39MEE866 pKa = 4.61LSSSRR871 pKa = 11.84RR872 pKa = 11.84WGQIVRR878 pKa = 11.84YY879 pKa = 8.48LRR881 pKa = 11.84RR882 pKa = 11.84IGRR885 pKa = 11.84VSGNDD890 pKa = 3.46GEE892 pKa = 4.54EE893 pKa = 3.97RR894 pKa = 11.84SSNEE898 pKa = 3.59SPATTKK904 pKa = 9.49TQGSAAAWGPPQEE917 pKa = 4.16AWPVDD922 pKa = 3.66GASLSTFEE930 pKa = 4.94PSSSGWFHH938 pKa = 7.76LEE940 pKa = 3.5GWW942 pKa = 3.69
Molecular weight: 103.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q89504|P2A_CFMVN Polyprotein P2A OS=Cocksfoot mottle virus (isolate Dactylis glomerata/Norway/CfMV-NO/1995) OX=1005059 GN=ORF2A PE=1 SV=2
MM1 pKa = 7.21MVRR4 pKa = 11.84KK5 pKa = 9.39GAATKK10 pKa = 10.44APQQPKK16 pKa = 10.04PKK18 pKa = 10.12AQQQPGGRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84GRR32 pKa = 11.84SMEE35 pKa = 4.1PVSRR39 pKa = 11.84PLNPPAAVGSTLKK52 pKa = 10.65AGRR55 pKa = 11.84GRR57 pKa = 11.84TAGVSDD63 pKa = 3.66WFDD66 pKa = 2.81TGMITSYY73 pKa = 11.07LGGFQRR79 pKa = 11.84TAGTTDD85 pKa = 3.09SQVFIVSPAALDD97 pKa = 3.63RR98 pKa = 11.84VGTIAKK104 pKa = 9.89AYY106 pKa = 9.43ALWRR110 pKa = 11.84PKK112 pKa = 9.87HH113 pKa = 5.18WEE115 pKa = 3.41IVYY118 pKa = 10.19LPRR121 pKa = 11.84CSTQTDD127 pKa = 3.86GSIEE131 pKa = 3.98MGFLLDD137 pKa = 4.08YY138 pKa = 10.97ADD140 pKa = 4.32SVPTNTRR147 pKa = 11.84TMASSTSFTTSNVWGGGDD165 pKa = 3.72GSSLLHH171 pKa = 6.06TSVKK175 pKa = 10.89SMGNAVTSALPCDD188 pKa = 3.66EE189 pKa = 5.84FSNKK193 pKa = 7.29WFKK196 pKa = 11.1LSWSTPEE203 pKa = 3.78EE204 pKa = 4.32SEE206 pKa = 4.28NAHH209 pKa = 5.97LTDD212 pKa = 3.53TYY214 pKa = 10.7VPARR218 pKa = 11.84FVVRR222 pKa = 11.84SDD224 pKa = 3.89FPVVTADD231 pKa = 3.94QPGHH235 pKa = 5.59LWLRR239 pKa = 11.84SRR241 pKa = 11.84ILLKK245 pKa = 10.9GSVSPSTNLL254 pKa = 3.32
MM1 pKa = 7.21MVRR4 pKa = 11.84KK5 pKa = 9.39GAATKK10 pKa = 10.44APQQPKK16 pKa = 10.04PKK18 pKa = 10.12AQQQPGGRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84GRR32 pKa = 11.84SMEE35 pKa = 4.1PVSRR39 pKa = 11.84PLNPPAAVGSTLKK52 pKa = 10.65AGRR55 pKa = 11.84GRR57 pKa = 11.84TAGVSDD63 pKa = 3.66WFDD66 pKa = 2.81TGMITSYY73 pKa = 11.07LGGFQRR79 pKa = 11.84TAGTTDD85 pKa = 3.09SQVFIVSPAALDD97 pKa = 3.63RR98 pKa = 11.84VGTIAKK104 pKa = 9.89AYY106 pKa = 9.43ALWRR110 pKa = 11.84PKK112 pKa = 9.87HH113 pKa = 5.18WEE115 pKa = 3.41IVYY118 pKa = 10.19LPRR121 pKa = 11.84CSTQTDD127 pKa = 3.86GSIEE131 pKa = 3.98MGFLLDD137 pKa = 4.08YY138 pKa = 10.97ADD140 pKa = 4.32SVPTNTRR147 pKa = 11.84TMASSTSFTTSNVWGGGDD165 pKa = 3.72GSSLLHH171 pKa = 6.06TSVKK175 pKa = 10.89SMGNAVTSALPCDD188 pKa = 3.66EE189 pKa = 5.84FSNKK193 pKa = 7.29WFKK196 pKa = 11.1LSWSTPEE203 pKa = 3.78EE204 pKa = 4.32SEE206 pKa = 4.28NAHH209 pKa = 5.97LTDD212 pKa = 3.53TYY214 pKa = 10.7VPARR218 pKa = 11.84FVVRR222 pKa = 11.84SDD224 pKa = 3.89FPVVTADD231 pKa = 3.94QPGHH235 pKa = 5.59LWLRR239 pKa = 11.84SRR241 pKa = 11.84ILLKK245 pKa = 10.9GSVSPSTNLL254 pKa = 3.32
Molecular weight: 27.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1872 |
108 |
942 |
468.0 |
51.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.013 ± 0.936 | 2.404 ± 0.501 |
4.274 ± 0.133 | 6.25 ± 0.963 |
2.671 ± 0.603 | 7.532 ± 0.266 |
1.389 ± 0.265 | 3.953 ± 0.358 |
4.594 ± 0.498 | 8.814 ± 0.821 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.03 ± 0.176 | 2.564 ± 0.091 |
6.143 ± 0.303 | 2.938 ± 0.176 |
6.517 ± 0.285 | 11.271 ± 0.841 |
6.09 ± 1.112 | 7.959 ± 0.372 |
2.35 ± 0.321 | 2.244 ± 0.063 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
