
Hubei diptera virus 5
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phasivirus; Dipteran phasivirus
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
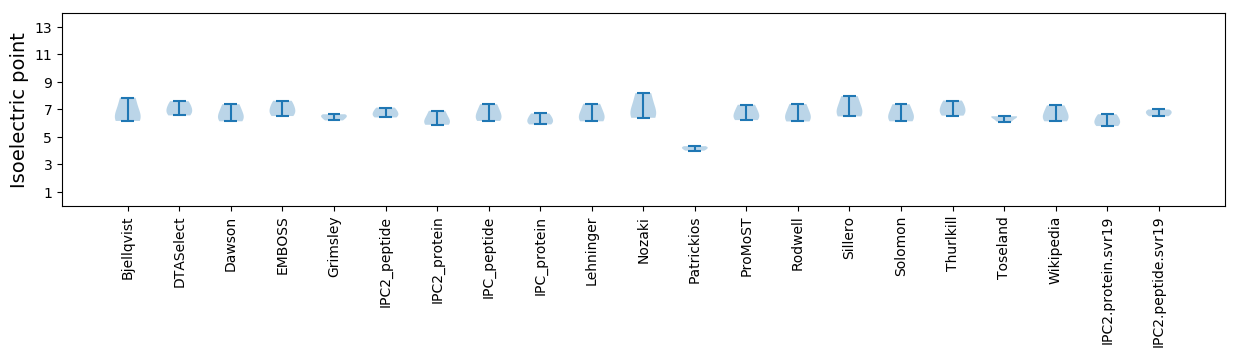
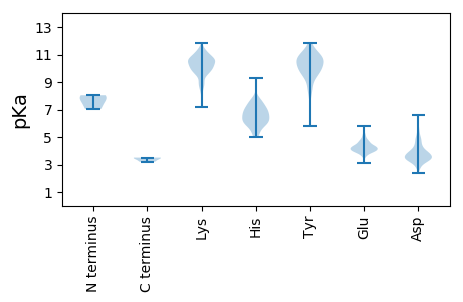
Protein with the lowest isoelectric point:
>tr|A0A1L3KPJ1|A0A1L3KPJ1_9VIRU Replicase OS=Hubei diptera virus 5 OX=1922886 PE=4 SV=1
MM1 pKa = 7.05YY2 pKa = 10.34RR3 pKa = 11.84FVDD6 pKa = 5.27GIKK9 pKa = 9.92EE10 pKa = 4.05YY11 pKa = 10.64LWSIVLNTTWVSAGLIKK28 pKa = 10.4DD29 pKa = 4.02YY30 pKa = 10.63QISWILDD37 pKa = 3.32QLKK40 pKa = 10.71DD41 pKa = 3.76KK42 pKa = 11.0EE43 pKa = 4.45LTTEE47 pKa = 4.71LLHH50 pKa = 6.69IDD52 pKa = 3.86CSSVQVFALFLYY64 pKa = 9.48DD65 pKa = 3.46VCLQVAKK72 pKa = 10.54VYY74 pKa = 10.76QLDD77 pKa = 4.04TEE79 pKa = 4.95LIRR82 pKa = 11.84YY83 pKa = 8.4PDD85 pKa = 4.75DD86 pKa = 3.7EE87 pKa = 4.46SADD90 pKa = 3.61YY91 pKa = 11.3SLARR95 pKa = 11.84LLTYY99 pKa = 9.61FYY101 pKa = 10.37THH103 pKa = 6.94LHH105 pKa = 6.84PILTVGLEE113 pKa = 4.32VEE115 pKa = 5.2DD116 pKa = 4.08STSTTGLVVQEE127 pKa = 4.77DD128 pKa = 4.31KK129 pKa = 11.16GTLTLFTGPSTSSRR143 pKa = 11.84IPVRR147 pKa = 11.84TSASKK152 pKa = 8.21PQQMSRR158 pKa = 11.84EE159 pKa = 4.14MLGLTDD165 pKa = 3.5WLRR168 pKa = 11.84PEE170 pKa = 4.27VATQAVTSEE179 pKa = 3.78VMAAYY184 pKa = 7.62EE185 pKa = 4.08TMEE188 pKa = 4.61KK189 pKa = 10.24SLCEE193 pKa = 4.05SGLTDD198 pKa = 3.63EE199 pKa = 5.6AIKK202 pKa = 9.6GTADD206 pKa = 3.02YY207 pKa = 9.94FAYY210 pKa = 10.72VGFNANIIRR219 pKa = 11.84AALVSCEE226 pKa = 3.44ADD228 pKa = 3.07EE229 pKa = 4.37RR230 pKa = 11.84KK231 pKa = 9.89RR232 pKa = 11.84SIEE235 pKa = 3.62ICTLIVTVLSRR246 pKa = 11.84GTSIAATGKK255 pKa = 10.69EE256 pKa = 4.07KK257 pKa = 10.91MNDD260 pKa = 2.93TAKK263 pKa = 11.29AMFATLCSKK272 pKa = 10.61YY273 pKa = 10.1RR274 pKa = 11.84IKK276 pKa = 10.42RR277 pKa = 11.84TLTAKK282 pKa = 10.28RR283 pKa = 11.84HH284 pKa = 5.58LGPNDD289 pKa = 3.21VTLSRR294 pKa = 11.84VVAAHH299 pKa = 6.51PEE301 pKa = 3.8IAYY304 pKa = 9.25KK305 pKa = 10.76ALLCRR310 pKa = 11.84DD311 pKa = 4.07LEE313 pKa = 4.28RR314 pKa = 11.84PIRR317 pKa = 11.84VQTMINTGFVDD328 pKa = 5.35FKK330 pKa = 11.23AGLRR334 pKa = 11.84GSYY337 pKa = 9.88VFSVVPTNQKK347 pKa = 10.21GIKK350 pKa = 9.59HH351 pKa = 6.03SGAAAAALTYY361 pKa = 10.05MAMEE365 pKa = 4.45TVLLRR370 pKa = 11.84DD371 pKa = 3.62KK372 pKa = 10.57NAPAITLEE380 pKa = 4.08EE381 pKa = 4.29ALTNCATFARR391 pKa = 11.84ATLNSSRR398 pKa = 11.84YY399 pKa = 10.26DD400 pKa = 3.73DD401 pKa = 3.79DD402 pKa = 3.83TKK404 pKa = 11.35KK405 pKa = 10.59RR406 pKa = 11.84WFDD409 pKa = 3.2KK410 pKa = 10.55MDD412 pKa = 3.52VQEE415 pKa = 4.39EE416 pKa = 4.25TAKK419 pKa = 10.48AWIRR423 pKa = 11.84RR424 pKa = 11.84FSAKK428 pKa = 10.04FPEE431 pKa = 4.72GLDD434 pKa = 3.57LLASLGFPTSVAA446 pKa = 3.48
MM1 pKa = 7.05YY2 pKa = 10.34RR3 pKa = 11.84FVDD6 pKa = 5.27GIKK9 pKa = 9.92EE10 pKa = 4.05YY11 pKa = 10.64LWSIVLNTTWVSAGLIKK28 pKa = 10.4DD29 pKa = 4.02YY30 pKa = 10.63QISWILDD37 pKa = 3.32QLKK40 pKa = 10.71DD41 pKa = 3.76KK42 pKa = 11.0EE43 pKa = 4.45LTTEE47 pKa = 4.71LLHH50 pKa = 6.69IDD52 pKa = 3.86CSSVQVFALFLYY64 pKa = 9.48DD65 pKa = 3.46VCLQVAKK72 pKa = 10.54VYY74 pKa = 10.76QLDD77 pKa = 4.04TEE79 pKa = 4.95LIRR82 pKa = 11.84YY83 pKa = 8.4PDD85 pKa = 4.75DD86 pKa = 3.7EE87 pKa = 4.46SADD90 pKa = 3.61YY91 pKa = 11.3SLARR95 pKa = 11.84LLTYY99 pKa = 9.61FYY101 pKa = 10.37THH103 pKa = 6.94LHH105 pKa = 6.84PILTVGLEE113 pKa = 4.32VEE115 pKa = 5.2DD116 pKa = 4.08STSTTGLVVQEE127 pKa = 4.77DD128 pKa = 4.31KK129 pKa = 11.16GTLTLFTGPSTSSRR143 pKa = 11.84IPVRR147 pKa = 11.84TSASKK152 pKa = 8.21PQQMSRR158 pKa = 11.84EE159 pKa = 4.14MLGLTDD165 pKa = 3.5WLRR168 pKa = 11.84PEE170 pKa = 4.27VATQAVTSEE179 pKa = 3.78VMAAYY184 pKa = 7.62EE185 pKa = 4.08TMEE188 pKa = 4.61KK189 pKa = 10.24SLCEE193 pKa = 4.05SGLTDD198 pKa = 3.63EE199 pKa = 5.6AIKK202 pKa = 9.6GTADD206 pKa = 3.02YY207 pKa = 9.94FAYY210 pKa = 10.72VGFNANIIRR219 pKa = 11.84AALVSCEE226 pKa = 3.44ADD228 pKa = 3.07EE229 pKa = 4.37RR230 pKa = 11.84KK231 pKa = 9.89RR232 pKa = 11.84SIEE235 pKa = 3.62ICTLIVTVLSRR246 pKa = 11.84GTSIAATGKK255 pKa = 10.69EE256 pKa = 4.07KK257 pKa = 10.91MNDD260 pKa = 2.93TAKK263 pKa = 11.29AMFATLCSKK272 pKa = 10.61YY273 pKa = 10.1RR274 pKa = 11.84IKK276 pKa = 10.42RR277 pKa = 11.84TLTAKK282 pKa = 10.28RR283 pKa = 11.84HH284 pKa = 5.58LGPNDD289 pKa = 3.21VTLSRR294 pKa = 11.84VVAAHH299 pKa = 6.51PEE301 pKa = 3.8IAYY304 pKa = 9.25KK305 pKa = 10.76ALLCRR310 pKa = 11.84DD311 pKa = 4.07LEE313 pKa = 4.28RR314 pKa = 11.84PIRR317 pKa = 11.84VQTMINTGFVDD328 pKa = 5.35FKK330 pKa = 11.23AGLRR334 pKa = 11.84GSYY337 pKa = 9.88VFSVVPTNQKK347 pKa = 10.21GIKK350 pKa = 9.59HH351 pKa = 6.03SGAAAAALTYY361 pKa = 10.05MAMEE365 pKa = 4.45TVLLRR370 pKa = 11.84DD371 pKa = 3.62KK372 pKa = 10.57NAPAITLEE380 pKa = 4.08EE381 pKa = 4.29ALTNCATFARR391 pKa = 11.84ATLNSSRR398 pKa = 11.84YY399 pKa = 10.26DD400 pKa = 3.73DD401 pKa = 3.79DD402 pKa = 3.83TKK404 pKa = 11.35KK405 pKa = 10.59RR406 pKa = 11.84WFDD409 pKa = 3.2KK410 pKa = 10.55MDD412 pKa = 3.52VQEE415 pKa = 4.39EE416 pKa = 4.25TAKK419 pKa = 10.48AWIRR423 pKa = 11.84RR424 pKa = 11.84FSAKK428 pKa = 10.04FPEE431 pKa = 4.72GLDD434 pKa = 3.57LLASLGFPTSVAA446 pKa = 3.48
Molecular weight: 49.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KPK7|A0A1L3KPK7_9VIRU Putative glycoprotein OS=Hubei diptera virus 5 OX=1922886 PE=4 SV=1
MM1 pKa = 7.8AKK3 pKa = 9.98PLYY6 pKa = 10.13FRR8 pKa = 11.84ILIILVCRR16 pKa = 11.84VLLLFAVANNTSFGDD31 pKa = 3.69GSKK34 pKa = 10.02ILNIIQGGNFQTLNNHH50 pKa = 6.15GGFHH54 pKa = 6.69VKK56 pKa = 9.54TEE58 pKa = 4.08SDD60 pKa = 3.88KK61 pKa = 11.45INCPVCNCNNTEE73 pKa = 4.04LVNVIKK79 pKa = 9.93TFFEE83 pKa = 4.93SNNSSFKK90 pKa = 10.31TLNITNRR97 pKa = 11.84ATEE100 pKa = 4.02LTSNYY105 pKa = 8.7TLIKK109 pKa = 9.11MIPPSTSGNNDD120 pKa = 3.81DD121 pKa = 3.91NPKK124 pKa = 10.83AEE126 pKa = 4.53VEE128 pKa = 4.18LLSNTLNNQNQGSFIPPIMSFLVNRR153 pKa = 11.84IVHH156 pKa = 6.0EE157 pKa = 4.33EE158 pKa = 4.14PIEE161 pKa = 4.13MKK163 pKa = 10.32VLRR166 pKa = 11.84YY167 pKa = 8.63QDD169 pKa = 4.14KK170 pKa = 10.09IGQISEE176 pKa = 4.2QIKK179 pKa = 10.59DD180 pKa = 3.64IIFEE184 pKa = 4.25DD185 pKa = 3.97TSIVCDD191 pKa = 3.59TKK193 pKa = 11.03SLKK196 pKa = 10.59KK197 pKa = 9.95MLLCVSTYY205 pKa = 10.25IGEE208 pKa = 4.45DD209 pKa = 2.73QDD211 pKa = 4.86KK212 pKa = 10.25NIKK215 pKa = 8.64VLSMLLNLISDD226 pKa = 4.72FLSEE230 pKa = 4.47FKK232 pKa = 10.88LRR234 pKa = 11.84NKK236 pKa = 9.63NNNYY240 pKa = 10.04DD241 pKa = 3.84GNVKK245 pKa = 10.19LGKK248 pKa = 9.85FSHH251 pKa = 6.93LMDD254 pKa = 4.84QNDD257 pKa = 3.44KK258 pKa = 11.35SNAISSFGRR267 pKa = 11.84KK268 pKa = 8.91LLSVNKK274 pKa = 10.05DD275 pKa = 3.25VLFTSEE281 pKa = 4.49VEE283 pKa = 4.11PGTSIMRR290 pKa = 11.84SKK292 pKa = 10.37PDD294 pKa = 3.18EE295 pKa = 3.95YY296 pKa = 10.93DD297 pKa = 2.86ISVRR301 pKa = 11.84LSRR304 pKa = 11.84IMEE307 pKa = 4.27GPKK310 pKa = 9.63TSNIDD315 pKa = 3.1AVKK318 pKa = 10.03AFEE321 pKa = 4.14IFRR324 pKa = 11.84VHH326 pKa = 7.6CDD328 pKa = 3.01RR329 pKa = 11.84KK330 pKa = 10.23HH331 pKa = 6.6NDD333 pKa = 2.81VNRR336 pKa = 11.84LDD338 pKa = 4.64EE339 pKa = 5.18IEE341 pKa = 4.57LSAIVKK347 pKa = 8.41PHH349 pKa = 6.81FSTTTQVCFSTFLCPTDD366 pKa = 3.35YY367 pKa = 11.25HH368 pKa = 7.6FDD370 pKa = 3.44FTTRR374 pKa = 11.84EE375 pKa = 4.35CKK377 pKa = 9.94QSLDD381 pKa = 3.4GALLNDD387 pKa = 3.55EE388 pKa = 5.32FDD390 pKa = 4.02FKK392 pKa = 11.13SIKK395 pKa = 10.13LSSIVGRR402 pKa = 11.84VTSHH406 pKa = 6.67PFEE409 pKa = 4.59TIIPRR414 pKa = 11.84LKK416 pKa = 10.65KK417 pKa = 10.39NYY419 pKa = 9.86CKK421 pKa = 9.73IDD423 pKa = 3.09NHH425 pKa = 6.77SIRR428 pKa = 11.84KK429 pKa = 8.64CVGKK433 pKa = 10.22IKK435 pKa = 10.61KK436 pKa = 9.56FEE438 pKa = 4.32KK439 pKa = 10.55DD440 pKa = 3.2FMAIHH445 pKa = 6.43VANDD449 pKa = 3.26WVLVDD454 pKa = 4.47GDD456 pKa = 4.25YY457 pKa = 11.45LLTMVQDD464 pKa = 4.02SKK466 pKa = 11.7DD467 pKa = 3.42FRR469 pKa = 11.84SYY471 pKa = 11.0VCNSSNPLIIPDD483 pKa = 3.9TDD485 pKa = 3.41CRR487 pKa = 11.84GDD489 pKa = 3.43DD490 pKa = 3.44SFIRR494 pKa = 11.84AFGRR498 pKa = 11.84GNSDD502 pKa = 3.34CFCRR506 pKa = 11.84INNNWHH512 pKa = 6.72DD513 pKa = 4.26LVFKK517 pKa = 10.99NLDD520 pKa = 3.32GVSYY524 pKa = 10.42IDD526 pKa = 3.26AWIAFTWTVEE536 pKa = 3.65IDD538 pKa = 3.6SKK540 pKa = 11.49VIEE543 pKa = 4.69NLNCDD548 pKa = 3.66DD549 pKa = 6.62CEE551 pKa = 4.44FKK553 pKa = 10.91CKK555 pKa = 10.58NGIIDD560 pKa = 3.91YY561 pKa = 9.96SVKK564 pKa = 10.53KK565 pKa = 10.45YY566 pKa = 10.3ISLMKK571 pKa = 9.87ICFMDD576 pKa = 3.31HH577 pKa = 7.08CILRR581 pKa = 11.84DD582 pKa = 3.45IKK584 pKa = 11.13QMSGSIRR591 pKa = 11.84IPEE594 pKa = 3.8LRR596 pKa = 11.84FYY598 pKa = 11.14GSKK601 pKa = 10.61FKK603 pKa = 9.71ITLLSEE609 pKa = 4.2TKK611 pKa = 8.9TGEE614 pKa = 4.08FITEE618 pKa = 4.07VDD620 pKa = 3.95CSNIDD625 pKa = 3.41LCQSISCVFCLEE637 pKa = 3.89RR638 pKa = 11.84LANYY642 pKa = 9.25HH643 pKa = 6.34CLSSYY648 pKa = 11.35LLILYY653 pKa = 8.45VLILFATFLILSIIYY668 pKa = 10.12KK669 pKa = 9.96IMKK672 pKa = 8.92LLKK675 pKa = 10.24AIFMSLFVTLKK686 pKa = 10.29FSYY689 pKa = 10.72KK690 pKa = 9.36LVRR693 pKa = 11.84YY694 pKa = 9.58ICIKK698 pKa = 9.91FYY700 pKa = 10.41RR701 pKa = 11.84YY702 pKa = 9.74CRR704 pKa = 11.84GHH706 pKa = 5.93MSRR709 pKa = 11.84SYY711 pKa = 11.14NRR713 pKa = 11.84VVEE716 pKa = 5.79LDD718 pKa = 4.69DD719 pKa = 3.56QDD721 pKa = 4.03LWEE724 pKa = 4.63TPRR727 pKa = 11.84QDD729 pKa = 3.68EE730 pKa = 4.23EE731 pKa = 4.94SMEE734 pKa = 4.07DD735 pKa = 3.72VRR737 pKa = 11.84IDD739 pKa = 3.36DD740 pKa = 4.29HH741 pKa = 9.32SPMNRR746 pKa = 11.84TPMRR750 pKa = 11.84RR751 pKa = 11.84PRR753 pKa = 11.84VLPANRR759 pKa = 11.84TSRR762 pKa = 11.84DD763 pKa = 2.73IKK765 pKa = 9.79MSLFIVATIITLAFSCSDD783 pKa = 2.91IGSISTQDD791 pKa = 3.48VSCTEE796 pKa = 3.89YY797 pKa = 11.08KK798 pKa = 9.6NTISCVTDD806 pKa = 3.55FNIMLTVAPMGQYY819 pKa = 11.18ACIQAKK825 pKa = 10.1SIEE828 pKa = 4.1GRR830 pKa = 11.84VIMSIRR836 pKa = 11.84IKK838 pKa = 9.83TKK840 pKa = 10.2SIKK843 pKa = 10.26LICNKK848 pKa = 9.84RR849 pKa = 11.84SLYY852 pKa = 8.17YY853 pKa = 9.94TYY855 pKa = 9.79RR856 pKa = 11.84TTPTISKK863 pKa = 9.98VYY865 pKa = 9.48RR866 pKa = 11.84CDD868 pKa = 3.66SAGEE872 pKa = 4.05CSKK875 pKa = 10.94NVCNSNDD882 pKa = 3.17PKK884 pKa = 10.82IFEE887 pKa = 4.76KK888 pKa = 10.68EE889 pKa = 4.0LNPNEE894 pKa = 4.32LKK896 pKa = 11.13VLGNTGCKK904 pKa = 9.51SVNGFWGNGCFYY916 pKa = 10.92ASSACIFYY924 pKa = 10.55RR925 pKa = 11.84VLFPNVEE932 pKa = 3.92NGDD935 pKa = 3.82FYY937 pKa = 11.84EE938 pKa = 5.14MFDD941 pKa = 3.8CPHH944 pKa = 7.14WSWQIEE950 pKa = 4.15LEE952 pKa = 4.2ITQSDD957 pKa = 5.1LIQEE961 pKa = 4.44KK962 pKa = 10.06VSSVVLSNEE971 pKa = 4.14LPTQSSIGSIRR982 pKa = 11.84LHH984 pKa = 5.99SVSVPPSPVLSDD996 pKa = 3.95CFMSSSIAGMNFISLTNCNKK1016 pKa = 9.79KK1017 pKa = 10.33DD1018 pKa = 3.55QLIKK1022 pKa = 11.2GEE1024 pKa = 4.4LGEE1027 pKa = 4.22IQCPSGVDD1035 pKa = 3.19AGNATNKK1042 pKa = 9.81CSYY1045 pKa = 10.1SSSIYY1050 pKa = 10.81SVTASGNDD1058 pKa = 3.02IQFASEE1064 pKa = 4.27VINPKK1069 pKa = 8.96DD1070 pKa = 3.19TSKK1073 pKa = 11.38AKK1075 pKa = 10.33RR1076 pKa = 11.84LPLNFDD1082 pKa = 3.57GFMLDD1087 pKa = 3.79IDD1089 pKa = 3.86NSIPIARR1096 pKa = 11.84MFGSSLFQFSIATSKK1111 pKa = 11.26YY1112 pKa = 8.61KK1113 pKa = 10.77FEE1115 pKa = 4.11ILKK1118 pKa = 10.06EE1119 pKa = 3.97RR1120 pKa = 11.84SSCSTTFLNLTGCYY1134 pKa = 9.56NCLGGATLCLKK1145 pKa = 10.52VDD1147 pKa = 3.44VDD1149 pKa = 4.31KK1150 pKa = 11.17EE1151 pKa = 4.29SSILKK1156 pKa = 9.69VDD1158 pKa = 3.87CPKK1161 pKa = 11.16SKK1163 pKa = 10.2ISTVFMVNKK1172 pKa = 10.09NSRR1175 pKa = 11.84EE1176 pKa = 4.09YY1177 pKa = 10.68CSKK1180 pKa = 10.74LSSNVQTIHH1189 pKa = 6.14EE1190 pKa = 4.36EE1191 pKa = 4.3CEE1193 pKa = 4.26IVCSGNTEE1201 pKa = 4.18KK1202 pKa = 11.13VKK1204 pKa = 11.23LEE1206 pKa = 3.95ASLSYY1211 pKa = 10.62DD1212 pKa = 3.16VNVIHH1217 pKa = 7.6KK1218 pKa = 10.52DD1219 pKa = 3.18NTNYY1223 pKa = 10.07IYY1225 pKa = 10.98NGVGILRR1232 pKa = 11.84GVDD1235 pKa = 2.85ISTISLWSLGFIILGFLIFLIIILLIKK1262 pKa = 10.38KK1263 pKa = 9.93CLFDD1267 pKa = 4.44HH1268 pKa = 6.29KK1269 pKa = 8.84TLKK1272 pKa = 10.99AFDD1275 pKa = 4.78PSSKK1279 pKa = 10.72SII1281 pKa = 3.51
MM1 pKa = 7.8AKK3 pKa = 9.98PLYY6 pKa = 10.13FRR8 pKa = 11.84ILIILVCRR16 pKa = 11.84VLLLFAVANNTSFGDD31 pKa = 3.69GSKK34 pKa = 10.02ILNIIQGGNFQTLNNHH50 pKa = 6.15GGFHH54 pKa = 6.69VKK56 pKa = 9.54TEE58 pKa = 4.08SDD60 pKa = 3.88KK61 pKa = 11.45INCPVCNCNNTEE73 pKa = 4.04LVNVIKK79 pKa = 9.93TFFEE83 pKa = 4.93SNNSSFKK90 pKa = 10.31TLNITNRR97 pKa = 11.84ATEE100 pKa = 4.02LTSNYY105 pKa = 8.7TLIKK109 pKa = 9.11MIPPSTSGNNDD120 pKa = 3.81DD121 pKa = 3.91NPKK124 pKa = 10.83AEE126 pKa = 4.53VEE128 pKa = 4.18LLSNTLNNQNQGSFIPPIMSFLVNRR153 pKa = 11.84IVHH156 pKa = 6.0EE157 pKa = 4.33EE158 pKa = 4.14PIEE161 pKa = 4.13MKK163 pKa = 10.32VLRR166 pKa = 11.84YY167 pKa = 8.63QDD169 pKa = 4.14KK170 pKa = 10.09IGQISEE176 pKa = 4.2QIKK179 pKa = 10.59DD180 pKa = 3.64IIFEE184 pKa = 4.25DD185 pKa = 3.97TSIVCDD191 pKa = 3.59TKK193 pKa = 11.03SLKK196 pKa = 10.59KK197 pKa = 9.95MLLCVSTYY205 pKa = 10.25IGEE208 pKa = 4.45DD209 pKa = 2.73QDD211 pKa = 4.86KK212 pKa = 10.25NIKK215 pKa = 8.64VLSMLLNLISDD226 pKa = 4.72FLSEE230 pKa = 4.47FKK232 pKa = 10.88LRR234 pKa = 11.84NKK236 pKa = 9.63NNNYY240 pKa = 10.04DD241 pKa = 3.84GNVKK245 pKa = 10.19LGKK248 pKa = 9.85FSHH251 pKa = 6.93LMDD254 pKa = 4.84QNDD257 pKa = 3.44KK258 pKa = 11.35SNAISSFGRR267 pKa = 11.84KK268 pKa = 8.91LLSVNKK274 pKa = 10.05DD275 pKa = 3.25VLFTSEE281 pKa = 4.49VEE283 pKa = 4.11PGTSIMRR290 pKa = 11.84SKK292 pKa = 10.37PDD294 pKa = 3.18EE295 pKa = 3.95YY296 pKa = 10.93DD297 pKa = 2.86ISVRR301 pKa = 11.84LSRR304 pKa = 11.84IMEE307 pKa = 4.27GPKK310 pKa = 9.63TSNIDD315 pKa = 3.1AVKK318 pKa = 10.03AFEE321 pKa = 4.14IFRR324 pKa = 11.84VHH326 pKa = 7.6CDD328 pKa = 3.01RR329 pKa = 11.84KK330 pKa = 10.23HH331 pKa = 6.6NDD333 pKa = 2.81VNRR336 pKa = 11.84LDD338 pKa = 4.64EE339 pKa = 5.18IEE341 pKa = 4.57LSAIVKK347 pKa = 8.41PHH349 pKa = 6.81FSTTTQVCFSTFLCPTDD366 pKa = 3.35YY367 pKa = 11.25HH368 pKa = 7.6FDD370 pKa = 3.44FTTRR374 pKa = 11.84EE375 pKa = 4.35CKK377 pKa = 9.94QSLDD381 pKa = 3.4GALLNDD387 pKa = 3.55EE388 pKa = 5.32FDD390 pKa = 4.02FKK392 pKa = 11.13SIKK395 pKa = 10.13LSSIVGRR402 pKa = 11.84VTSHH406 pKa = 6.67PFEE409 pKa = 4.59TIIPRR414 pKa = 11.84LKK416 pKa = 10.65KK417 pKa = 10.39NYY419 pKa = 9.86CKK421 pKa = 9.73IDD423 pKa = 3.09NHH425 pKa = 6.77SIRR428 pKa = 11.84KK429 pKa = 8.64CVGKK433 pKa = 10.22IKK435 pKa = 10.61KK436 pKa = 9.56FEE438 pKa = 4.32KK439 pKa = 10.55DD440 pKa = 3.2FMAIHH445 pKa = 6.43VANDD449 pKa = 3.26WVLVDD454 pKa = 4.47GDD456 pKa = 4.25YY457 pKa = 11.45LLTMVQDD464 pKa = 4.02SKK466 pKa = 11.7DD467 pKa = 3.42FRR469 pKa = 11.84SYY471 pKa = 11.0VCNSSNPLIIPDD483 pKa = 3.9TDD485 pKa = 3.41CRR487 pKa = 11.84GDD489 pKa = 3.43DD490 pKa = 3.44SFIRR494 pKa = 11.84AFGRR498 pKa = 11.84GNSDD502 pKa = 3.34CFCRR506 pKa = 11.84INNNWHH512 pKa = 6.72DD513 pKa = 4.26LVFKK517 pKa = 10.99NLDD520 pKa = 3.32GVSYY524 pKa = 10.42IDD526 pKa = 3.26AWIAFTWTVEE536 pKa = 3.65IDD538 pKa = 3.6SKK540 pKa = 11.49VIEE543 pKa = 4.69NLNCDD548 pKa = 3.66DD549 pKa = 6.62CEE551 pKa = 4.44FKK553 pKa = 10.91CKK555 pKa = 10.58NGIIDD560 pKa = 3.91YY561 pKa = 9.96SVKK564 pKa = 10.53KK565 pKa = 10.45YY566 pKa = 10.3ISLMKK571 pKa = 9.87ICFMDD576 pKa = 3.31HH577 pKa = 7.08CILRR581 pKa = 11.84DD582 pKa = 3.45IKK584 pKa = 11.13QMSGSIRR591 pKa = 11.84IPEE594 pKa = 3.8LRR596 pKa = 11.84FYY598 pKa = 11.14GSKK601 pKa = 10.61FKK603 pKa = 9.71ITLLSEE609 pKa = 4.2TKK611 pKa = 8.9TGEE614 pKa = 4.08FITEE618 pKa = 4.07VDD620 pKa = 3.95CSNIDD625 pKa = 3.41LCQSISCVFCLEE637 pKa = 3.89RR638 pKa = 11.84LANYY642 pKa = 9.25HH643 pKa = 6.34CLSSYY648 pKa = 11.35LLILYY653 pKa = 8.45VLILFATFLILSIIYY668 pKa = 10.12KK669 pKa = 9.96IMKK672 pKa = 8.92LLKK675 pKa = 10.24AIFMSLFVTLKK686 pKa = 10.29FSYY689 pKa = 10.72KK690 pKa = 9.36LVRR693 pKa = 11.84YY694 pKa = 9.58ICIKK698 pKa = 9.91FYY700 pKa = 10.41RR701 pKa = 11.84YY702 pKa = 9.74CRR704 pKa = 11.84GHH706 pKa = 5.93MSRR709 pKa = 11.84SYY711 pKa = 11.14NRR713 pKa = 11.84VVEE716 pKa = 5.79LDD718 pKa = 4.69DD719 pKa = 3.56QDD721 pKa = 4.03LWEE724 pKa = 4.63TPRR727 pKa = 11.84QDD729 pKa = 3.68EE730 pKa = 4.23EE731 pKa = 4.94SMEE734 pKa = 4.07DD735 pKa = 3.72VRR737 pKa = 11.84IDD739 pKa = 3.36DD740 pKa = 4.29HH741 pKa = 9.32SPMNRR746 pKa = 11.84TPMRR750 pKa = 11.84RR751 pKa = 11.84PRR753 pKa = 11.84VLPANRR759 pKa = 11.84TSRR762 pKa = 11.84DD763 pKa = 2.73IKK765 pKa = 9.79MSLFIVATIITLAFSCSDD783 pKa = 2.91IGSISTQDD791 pKa = 3.48VSCTEE796 pKa = 3.89YY797 pKa = 11.08KK798 pKa = 9.6NTISCVTDD806 pKa = 3.55FNIMLTVAPMGQYY819 pKa = 11.18ACIQAKK825 pKa = 10.1SIEE828 pKa = 4.1GRR830 pKa = 11.84VIMSIRR836 pKa = 11.84IKK838 pKa = 9.83TKK840 pKa = 10.2SIKK843 pKa = 10.26LICNKK848 pKa = 9.84RR849 pKa = 11.84SLYY852 pKa = 8.17YY853 pKa = 9.94TYY855 pKa = 9.79RR856 pKa = 11.84TTPTISKK863 pKa = 9.98VYY865 pKa = 9.48RR866 pKa = 11.84CDD868 pKa = 3.66SAGEE872 pKa = 4.05CSKK875 pKa = 10.94NVCNSNDD882 pKa = 3.17PKK884 pKa = 10.82IFEE887 pKa = 4.76KK888 pKa = 10.68EE889 pKa = 4.0LNPNEE894 pKa = 4.32LKK896 pKa = 11.13VLGNTGCKK904 pKa = 9.51SVNGFWGNGCFYY916 pKa = 10.92ASSACIFYY924 pKa = 10.55RR925 pKa = 11.84VLFPNVEE932 pKa = 3.92NGDD935 pKa = 3.82FYY937 pKa = 11.84EE938 pKa = 5.14MFDD941 pKa = 3.8CPHH944 pKa = 7.14WSWQIEE950 pKa = 4.15LEE952 pKa = 4.2ITQSDD957 pKa = 5.1LIQEE961 pKa = 4.44KK962 pKa = 10.06VSSVVLSNEE971 pKa = 4.14LPTQSSIGSIRR982 pKa = 11.84LHH984 pKa = 5.99SVSVPPSPVLSDD996 pKa = 3.95CFMSSSIAGMNFISLTNCNKK1016 pKa = 9.79KK1017 pKa = 10.33DD1018 pKa = 3.55QLIKK1022 pKa = 11.2GEE1024 pKa = 4.4LGEE1027 pKa = 4.22IQCPSGVDD1035 pKa = 3.19AGNATNKK1042 pKa = 9.81CSYY1045 pKa = 10.1SSSIYY1050 pKa = 10.81SVTASGNDD1058 pKa = 3.02IQFASEE1064 pKa = 4.27VINPKK1069 pKa = 8.96DD1070 pKa = 3.19TSKK1073 pKa = 11.38AKK1075 pKa = 10.33RR1076 pKa = 11.84LPLNFDD1082 pKa = 3.57GFMLDD1087 pKa = 3.79IDD1089 pKa = 3.86NSIPIARR1096 pKa = 11.84MFGSSLFQFSIATSKK1111 pKa = 11.26YY1112 pKa = 8.61KK1113 pKa = 10.77FEE1115 pKa = 4.11ILKK1118 pKa = 10.06EE1119 pKa = 3.97RR1120 pKa = 11.84SSCSTTFLNLTGCYY1134 pKa = 9.56NCLGGATLCLKK1145 pKa = 10.52VDD1147 pKa = 3.44VDD1149 pKa = 4.31KK1150 pKa = 11.17EE1151 pKa = 4.29SSILKK1156 pKa = 9.69VDD1158 pKa = 3.87CPKK1161 pKa = 11.16SKK1163 pKa = 10.2ISTVFMVNKK1172 pKa = 10.09NSRR1175 pKa = 11.84EE1176 pKa = 4.09YY1177 pKa = 10.68CSKK1180 pKa = 10.74LSSNVQTIHH1189 pKa = 6.14EE1190 pKa = 4.36EE1191 pKa = 4.3CEE1193 pKa = 4.26IVCSGNTEE1201 pKa = 4.18KK1202 pKa = 11.13VKK1204 pKa = 11.23LEE1206 pKa = 3.95ASLSYY1211 pKa = 10.62DD1212 pKa = 3.16VNVIHH1217 pKa = 7.6KK1218 pKa = 10.52DD1219 pKa = 3.18NTNYY1223 pKa = 10.07IYY1225 pKa = 10.98NGVGILRR1232 pKa = 11.84GVDD1235 pKa = 2.85ISTISLWSLGFIILGFLIFLIIILLIKK1262 pKa = 10.38KK1263 pKa = 9.93CLFDD1267 pKa = 4.44HH1268 pKa = 6.29KK1269 pKa = 8.84TLKK1272 pKa = 10.99AFDD1275 pKa = 4.78PSSKK1279 pKa = 10.72SII1281 pKa = 3.51
Molecular weight: 145.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3983 |
446 |
2256 |
1327.7 |
152.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.866 ± 1.72 | 2.234 ± 0.781 |
5.975 ± 0.154 | 6.528 ± 0.793 |
5.222 ± 0.453 | 4.168 ± 0.14 |
1.833 ± 0.179 | 8.285 ± 0.91 |
7.658 ± 0.545 | 9.691 ± 0.412 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.812 ± 0.244 | 5.674 ± 0.988 |
3.163 ± 0.097 | 2.611 ± 0.175 |
4.62 ± 0.335 | 9.515 ± 0.684 |
5.85 ± 1.053 | 5.523 ± 0.524 |
1.054 ± 0.15 | 3.716 ± 0.245 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
