
Agrococcus pavilionensis RW1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Agrococcus; Agrococcus pavilionensis
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
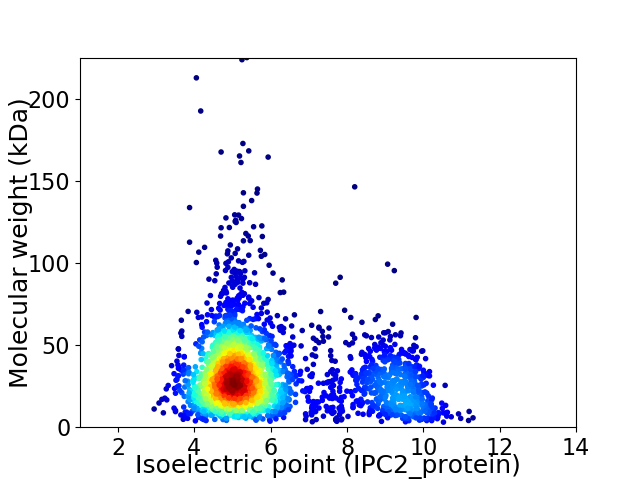
Virtual 2D-PAGE plot for 2776 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
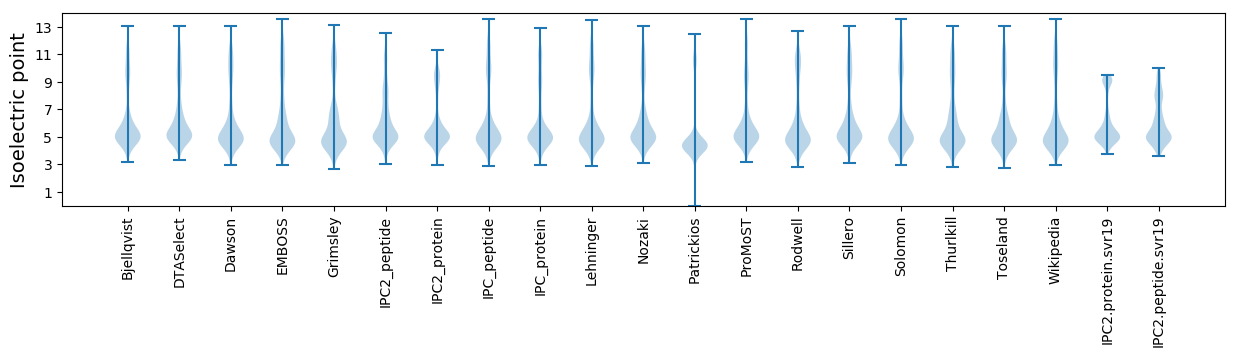
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U1LP10|U1LP10_9MICO YCII domain-containing protein OS=Agrococcus pavilionensis RW1 OX=1330458 GN=L332_06755 PE=3 SV=1
MM1 pKa = 7.75KK2 pKa = 9.14FTKK5 pKa = 10.26LGRR8 pKa = 11.84AAVVAAAGALLLSSCAANEE27 pKa = 4.17SGADD31 pKa = 3.32EE32 pKa = 4.48AAPEE36 pKa = 4.57GSQAAGSSLSGDD48 pKa = 3.54LAGAGASSMGAAQEE62 pKa = 3.7AWIAGFQTEE71 pKa = 4.28NPDD74 pKa = 3.33VNVTYY79 pKa = 10.73DD80 pKa = 3.62PAGSGAGRR88 pKa = 11.84EE89 pKa = 3.91QFIAGGVSFAGSDD102 pKa = 3.23AYY104 pKa = 11.28LKK106 pKa = 10.88DD107 pKa = 3.7EE108 pKa = 4.21EE109 pKa = 4.97LEE111 pKa = 4.42GEE113 pKa = 4.62FGACVPDD120 pKa = 4.09TLPVDD125 pKa = 4.04LPVYY129 pKa = 10.01ISPIAVIFNVEE140 pKa = 4.33GVDD143 pKa = 4.07SLNMDD148 pKa = 3.97ASTIAKK154 pKa = 9.4IFSGQITSWDD164 pKa = 3.56DD165 pKa = 3.13PAIAEE170 pKa = 4.13QNEE173 pKa = 4.59GVEE176 pKa = 4.76LPATPIQPVHH186 pKa = 6.82RR187 pKa = 11.84SDD189 pKa = 5.61DD190 pKa = 3.57SGTTEE195 pKa = 4.6NFTEE199 pKa = 4.28YY200 pKa = 10.76LAATAAADD208 pKa = 3.25WTEE211 pKa = 4.32GVFEE215 pKa = 4.24TWPSSVYY222 pKa = 9.39STSEE226 pKa = 3.97GAQGTSGVVDD236 pKa = 3.88AVSNGTGTIGYY247 pKa = 9.82ADD249 pKa = 4.09ASRR252 pKa = 11.84AGDD255 pKa = 3.84LGTVALKK262 pKa = 10.81VGEE265 pKa = 4.28EE266 pKa = 4.18YY267 pKa = 11.43VPFTADD273 pKa = 2.88AAAAAVDD280 pKa = 3.58ASPRR284 pKa = 11.84VEE286 pKa = 4.08GRR288 pKa = 11.84HH289 pKa = 5.63EE290 pKa = 4.02GDD292 pKa = 2.97IAVEE296 pKa = 4.21LDD298 pKa = 3.18RR299 pKa = 11.84TTTEE303 pKa = 3.66AGVYY307 pKa = 8.98PLVLISYY314 pKa = 9.68AIACSEE320 pKa = 4.16YY321 pKa = 9.93QDD323 pKa = 4.6AEE325 pKa = 3.96AGEE328 pKa = 4.31LVKK331 pKa = 10.75AYY333 pKa = 10.34LSYY336 pKa = 10.75VASEE340 pKa = 4.28EE341 pKa = 4.17GQAAGQEE348 pKa = 4.36NAGNAPISDD357 pKa = 3.97EE358 pKa = 4.24LRR360 pKa = 11.84DD361 pKa = 3.9EE362 pKa = 4.14ILAVVDD368 pKa = 4.98SIQQ371 pKa = 3.27
MM1 pKa = 7.75KK2 pKa = 9.14FTKK5 pKa = 10.26LGRR8 pKa = 11.84AAVVAAAGALLLSSCAANEE27 pKa = 4.17SGADD31 pKa = 3.32EE32 pKa = 4.48AAPEE36 pKa = 4.57GSQAAGSSLSGDD48 pKa = 3.54LAGAGASSMGAAQEE62 pKa = 3.7AWIAGFQTEE71 pKa = 4.28NPDD74 pKa = 3.33VNVTYY79 pKa = 10.73DD80 pKa = 3.62PAGSGAGRR88 pKa = 11.84EE89 pKa = 3.91QFIAGGVSFAGSDD102 pKa = 3.23AYY104 pKa = 11.28LKK106 pKa = 10.88DD107 pKa = 3.7EE108 pKa = 4.21EE109 pKa = 4.97LEE111 pKa = 4.42GEE113 pKa = 4.62FGACVPDD120 pKa = 4.09TLPVDD125 pKa = 4.04LPVYY129 pKa = 10.01ISPIAVIFNVEE140 pKa = 4.33GVDD143 pKa = 4.07SLNMDD148 pKa = 3.97ASTIAKK154 pKa = 9.4IFSGQITSWDD164 pKa = 3.56DD165 pKa = 3.13PAIAEE170 pKa = 4.13QNEE173 pKa = 4.59GVEE176 pKa = 4.76LPATPIQPVHH186 pKa = 6.82RR187 pKa = 11.84SDD189 pKa = 5.61DD190 pKa = 3.57SGTTEE195 pKa = 4.6NFTEE199 pKa = 4.28YY200 pKa = 10.76LAATAAADD208 pKa = 3.25WTEE211 pKa = 4.32GVFEE215 pKa = 4.24TWPSSVYY222 pKa = 9.39STSEE226 pKa = 3.97GAQGTSGVVDD236 pKa = 3.88AVSNGTGTIGYY247 pKa = 9.82ADD249 pKa = 4.09ASRR252 pKa = 11.84AGDD255 pKa = 3.84LGTVALKK262 pKa = 10.81VGEE265 pKa = 4.28EE266 pKa = 4.18YY267 pKa = 11.43VPFTADD273 pKa = 2.88AAAAAVDD280 pKa = 3.58ASPRR284 pKa = 11.84VEE286 pKa = 4.08GRR288 pKa = 11.84HH289 pKa = 5.63EE290 pKa = 4.02GDD292 pKa = 2.97IAVEE296 pKa = 4.21LDD298 pKa = 3.18RR299 pKa = 11.84TTTEE303 pKa = 3.66AGVYY307 pKa = 8.98PLVLISYY314 pKa = 9.68AIACSEE320 pKa = 4.16YY321 pKa = 9.93QDD323 pKa = 4.6AEE325 pKa = 3.96AGEE328 pKa = 4.31LVKK331 pKa = 10.75AYY333 pKa = 10.34LSYY336 pKa = 10.75VASEE340 pKa = 4.28EE341 pKa = 4.17GQAAGQEE348 pKa = 4.36NAGNAPISDD357 pKa = 3.97EE358 pKa = 4.24LRR360 pKa = 11.84DD361 pKa = 3.9EE362 pKa = 4.14ILAVVDD368 pKa = 4.98SIQQ371 pKa = 3.27
Molecular weight: 37.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U1MNN2|U1MNN2_9MICO Uncharacterized protein OS=Agrococcus pavilionensis RW1 OX=1330458 GN=L332_03425 PE=4 SV=1
MM1 pKa = 7.56NILSRR6 pKa = 11.84LRR8 pKa = 11.84SAVSGRR14 pKa = 11.84RR15 pKa = 11.84TGGAARR21 pKa = 11.84GRR23 pKa = 11.84RR24 pKa = 11.84STGMGGAAGARR35 pKa = 11.84GRR37 pKa = 11.84GSLLSSALGWISGRR51 pKa = 11.84NRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 3.11
MM1 pKa = 7.56NILSRR6 pKa = 11.84LRR8 pKa = 11.84SAVSGRR14 pKa = 11.84RR15 pKa = 11.84TGGAARR21 pKa = 11.84GRR23 pKa = 11.84RR24 pKa = 11.84STGMGGAAGARR35 pKa = 11.84GRR37 pKa = 11.84GSLLSSALGWISGRR51 pKa = 11.84NRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 3.11
Molecular weight: 5.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
872884 |
29 |
2082 |
314.4 |
33.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.464 ± 0.067 | 0.475 ± 0.011 |
6.118 ± 0.044 | 6.472 ± 0.045 |
2.81 ± 0.03 | 9.062 ± 0.044 |
2.094 ± 0.023 | 4.343 ± 0.034 |
1.688 ± 0.026 | 10.343 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.9 ± 0.019 | 1.5 ± 0.023 |
5.43 ± 0.036 | 2.843 ± 0.025 |
8.008 ± 0.061 | 5.193 ± 0.031 |
5.286 ± 0.039 | 8.826 ± 0.044 |
1.506 ± 0.022 | 1.639 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
