
Roseovarius Plymouth podovirus 1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Schitoviridae; Rhodovirinae; Plymouthvirus
Average proteome isoelectric point is 5.71
Get precalculated fractions of proteins
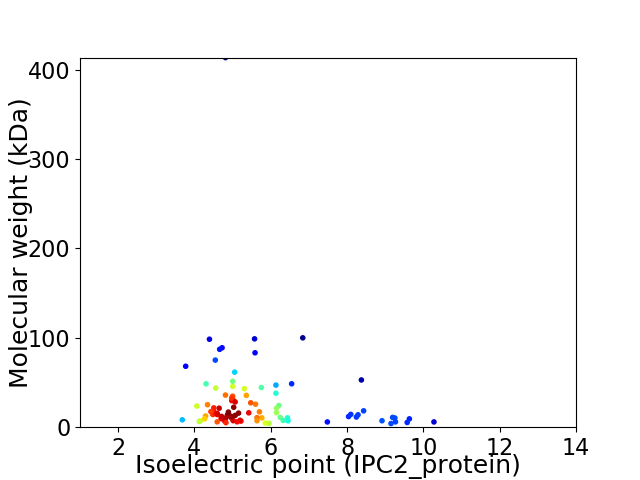
Virtual 2D-PAGE plot for 91 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4Q596|K4Q596_9CAUD Hypothetical host-like thioredoxin OS=Roseovarius Plymouth podovirus 1 OX=926474 PE=4 SV=1
MM1 pKa = 7.72NDD3 pKa = 3.3LNLTAVMEE11 pKa = 4.2PRR13 pKa = 11.84YY14 pKa = 10.49VLTIQDD20 pKa = 3.27QSVVVGSDD28 pKa = 3.47FNLTLQEE35 pKa = 3.73NSAPLKK41 pKa = 9.95LTLGLQGQKK50 pKa = 10.52GIQGDD55 pKa = 4.06VGPQGPVGPAGPAGTDD71 pKa = 3.28GTDD74 pKa = 3.27GVSFAFDD81 pKa = 3.43GTDD84 pKa = 2.83TFANRR89 pKa = 11.84NLYY92 pKa = 9.81DD93 pKa = 3.8AEE95 pKa = 4.42VVGWRR100 pKa = 11.84FLASDD105 pKa = 3.18TGEE108 pKa = 3.68IYY110 pKa = 10.93VRR112 pKa = 11.84LDD114 pKa = 3.13PTGWSAPIQLKK125 pKa = 10.75GDD127 pKa = 3.77TGPQGPQGIQGVQGPIGPQGNPGADD152 pKa = 3.16GQGFTVDD159 pKa = 4.43AYY161 pKa = 11.01AEE163 pKa = 4.13LVNLGTYY170 pKa = 10.61DD171 pKa = 3.53NEE173 pKa = 4.37AEE175 pKa = 4.34GFSVVDD181 pKa = 3.54PVTGLLYY188 pKa = 10.09IRR190 pKa = 11.84YY191 pKa = 8.73SVAGTWTNGIQFVGPEE207 pKa = 4.54GPTGPIGPAGPTGAPGAAGADD228 pKa = 3.59GADD231 pKa = 3.46FAFDD235 pKa = 3.62AVGTLPEE242 pKa = 4.36KK243 pKa = 10.69NLYY246 pKa = 8.54NAQPEE251 pKa = 4.68GFTFLDD257 pKa = 3.62EE258 pKa = 4.55VNGNFYY264 pKa = 10.71VRR266 pKa = 11.84NDD268 pKa = 3.29ATIGAWTNAIPFQGPTGPQGAQGDD292 pKa = 4.35AGISAYY298 pKa = 10.5DD299 pKa = 3.8LAVSQQGFVGSVSDD313 pKa = 3.5WLTTLTAYY321 pKa = 10.01GVAVSNGFVGTQADD335 pKa = 3.84WLIHH339 pKa = 5.27INAYY343 pKa = 9.9GIAVQNGYY351 pKa = 10.62LGTQAQWLQDD361 pKa = 3.28MTAYY365 pKa = 10.39GIAVLNGYY373 pKa = 10.36AGTEE377 pKa = 4.2PEE379 pKa = 4.09WLIHH383 pKa = 5.28INAYY387 pKa = 9.51GRR389 pKa = 11.84ALDD392 pKa = 3.63NGFVGTEE399 pKa = 4.1LEE401 pKa = 4.28WLDD404 pKa = 3.57SLTAYY409 pKa = 10.04GVAQSNGYY417 pKa = 10.58AGTKK421 pKa = 9.65AAWLTSLTAYY431 pKa = 9.88GVAVDD436 pKa = 3.79NGFVGTEE443 pKa = 4.22PEE445 pKa = 4.17WLTHH449 pKa = 5.25ISAYY453 pKa = 9.82GVAVQNGFVGTEE465 pKa = 4.04LQWLDD470 pKa = 3.89SLVGPIGPQGIQGLKK485 pKa = 10.33GDD487 pKa = 4.51TGQSLIPDD495 pKa = 5.31AIDD498 pKa = 3.87LFANRR503 pKa = 11.84ALYY506 pKa = 8.54DD507 pKa = 3.6TEE509 pKa = 5.35LEE511 pKa = 4.34GFVFMASDD519 pKa = 3.17TGLIYY524 pKa = 10.63FRR526 pKa = 11.84EE527 pKa = 4.4TVFNGTWSVGVPFRR541 pKa = 11.84GPDD544 pKa = 3.68GLPGDD549 pKa = 3.78VTTRR553 pKa = 11.84SLINTATYY561 pKa = 9.25TVLNTDD567 pKa = 3.68LQTGRR572 pKa = 11.84EE573 pKa = 4.02IKK575 pKa = 10.55KK576 pKa = 9.06VDD578 pKa = 3.58HH579 pKa = 6.84AVGCAITVPAGLTNQEE595 pKa = 4.17PCTFIQTNVGQITFAGAVGVTILSADD621 pKa = 3.44SALLSRR627 pKa = 11.84VQGSFVTLLPDD638 pKa = 4.11GDD640 pKa = 4.24TADD643 pKa = 4.47LYY645 pKa = 11.51YY646 pKa = 10.31LTGDD650 pKa = 3.4IVAA653 pKa = 4.9
MM1 pKa = 7.72NDD3 pKa = 3.3LNLTAVMEE11 pKa = 4.2PRR13 pKa = 11.84YY14 pKa = 10.49VLTIQDD20 pKa = 3.27QSVVVGSDD28 pKa = 3.47FNLTLQEE35 pKa = 3.73NSAPLKK41 pKa = 9.95LTLGLQGQKK50 pKa = 10.52GIQGDD55 pKa = 4.06VGPQGPVGPAGPAGTDD71 pKa = 3.28GTDD74 pKa = 3.27GVSFAFDD81 pKa = 3.43GTDD84 pKa = 2.83TFANRR89 pKa = 11.84NLYY92 pKa = 9.81DD93 pKa = 3.8AEE95 pKa = 4.42VVGWRR100 pKa = 11.84FLASDD105 pKa = 3.18TGEE108 pKa = 3.68IYY110 pKa = 10.93VRR112 pKa = 11.84LDD114 pKa = 3.13PTGWSAPIQLKK125 pKa = 10.75GDD127 pKa = 3.77TGPQGPQGIQGVQGPIGPQGNPGADD152 pKa = 3.16GQGFTVDD159 pKa = 4.43AYY161 pKa = 11.01AEE163 pKa = 4.13LVNLGTYY170 pKa = 10.61DD171 pKa = 3.53NEE173 pKa = 4.37AEE175 pKa = 4.34GFSVVDD181 pKa = 3.54PVTGLLYY188 pKa = 10.09IRR190 pKa = 11.84YY191 pKa = 8.73SVAGTWTNGIQFVGPEE207 pKa = 4.54GPTGPIGPAGPTGAPGAAGADD228 pKa = 3.59GADD231 pKa = 3.46FAFDD235 pKa = 3.62AVGTLPEE242 pKa = 4.36KK243 pKa = 10.69NLYY246 pKa = 8.54NAQPEE251 pKa = 4.68GFTFLDD257 pKa = 3.62EE258 pKa = 4.55VNGNFYY264 pKa = 10.71VRR266 pKa = 11.84NDD268 pKa = 3.29ATIGAWTNAIPFQGPTGPQGAQGDD292 pKa = 4.35AGISAYY298 pKa = 10.5DD299 pKa = 3.8LAVSQQGFVGSVSDD313 pKa = 3.5WLTTLTAYY321 pKa = 10.01GVAVSNGFVGTQADD335 pKa = 3.84WLIHH339 pKa = 5.27INAYY343 pKa = 9.9GIAVQNGYY351 pKa = 10.62LGTQAQWLQDD361 pKa = 3.28MTAYY365 pKa = 10.39GIAVLNGYY373 pKa = 10.36AGTEE377 pKa = 4.2PEE379 pKa = 4.09WLIHH383 pKa = 5.28INAYY387 pKa = 9.51GRR389 pKa = 11.84ALDD392 pKa = 3.63NGFVGTEE399 pKa = 4.1LEE401 pKa = 4.28WLDD404 pKa = 3.57SLTAYY409 pKa = 10.04GVAQSNGYY417 pKa = 10.58AGTKK421 pKa = 9.65AAWLTSLTAYY431 pKa = 9.88GVAVDD436 pKa = 3.79NGFVGTEE443 pKa = 4.22PEE445 pKa = 4.17WLTHH449 pKa = 5.25ISAYY453 pKa = 9.82GVAVQNGFVGTEE465 pKa = 4.04LQWLDD470 pKa = 3.89SLVGPIGPQGIQGLKK485 pKa = 10.33GDD487 pKa = 4.51TGQSLIPDD495 pKa = 5.31AIDD498 pKa = 3.87LFANRR503 pKa = 11.84ALYY506 pKa = 8.54DD507 pKa = 3.6TEE509 pKa = 5.35LEE511 pKa = 4.34GFVFMASDD519 pKa = 3.17TGLIYY524 pKa = 10.63FRR526 pKa = 11.84EE527 pKa = 4.4TVFNGTWSVGVPFRR541 pKa = 11.84GPDD544 pKa = 3.68GLPGDD549 pKa = 3.78VTTRR553 pKa = 11.84SLINTATYY561 pKa = 9.25TVLNTDD567 pKa = 3.68LQTGRR572 pKa = 11.84EE573 pKa = 4.02IKK575 pKa = 10.55KK576 pKa = 9.06VDD578 pKa = 3.58HH579 pKa = 6.84AVGCAITVPAGLTNQEE595 pKa = 4.17PCTFIQTNVGQITFAGAVGVTILSADD621 pKa = 3.44SALLSRR627 pKa = 11.84VQGSFVTLLPDD638 pKa = 4.11GDD640 pKa = 4.24TADD643 pKa = 4.47LYY645 pKa = 11.51YY646 pKa = 10.31LTGDD650 pKa = 3.4IVAA653 pKa = 4.9
Molecular weight: 68.18 kDa
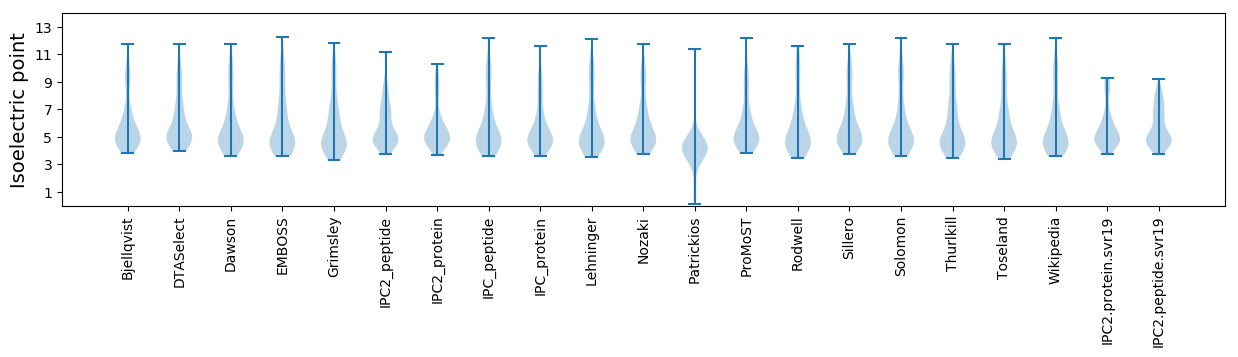
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4Q4U2|K4Q4U2_9CAUD N4 gp24-like protein OS=Roseovarius Plymouth podovirus 1 OX=926474 PE=4 SV=1
MM1 pKa = 7.99EE2 pKa = 4.5IRR4 pKa = 11.84EE5 pKa = 3.97YY6 pKa = 10.96RR7 pKa = 11.84QFAEE11 pKa = 4.73ALMSNCYY18 pKa = 10.04LNRR21 pKa = 11.84EE22 pKa = 4.12AQPCKK27 pKa = 10.52LKK29 pKa = 10.63NAKK32 pKa = 8.27TKK34 pKa = 10.3KK35 pKa = 10.07LKK37 pKa = 10.18NLRR40 pKa = 11.84SGIRR44 pKa = 11.84KK45 pKa = 9.13SPYY48 pKa = 9.4LLKK51 pKa = 10.83SIIWKK56 pKa = 8.8EE57 pKa = 3.64QLRR60 pKa = 11.84QIEE63 pKa = 4.13QEE65 pKa = 3.88LAKK68 pKa = 10.24RR69 pKa = 11.84IHH71 pKa = 6.2NGRR74 pKa = 11.84RR75 pKa = 11.84YY76 pKa = 9.27PP77 pKa = 3.53
MM1 pKa = 7.99EE2 pKa = 4.5IRR4 pKa = 11.84EE5 pKa = 3.97YY6 pKa = 10.96RR7 pKa = 11.84QFAEE11 pKa = 4.73ALMSNCYY18 pKa = 10.04LNRR21 pKa = 11.84EE22 pKa = 4.12AQPCKK27 pKa = 10.52LKK29 pKa = 10.63NAKK32 pKa = 8.27TKK34 pKa = 10.3KK35 pKa = 10.07LKK37 pKa = 10.18NLRR40 pKa = 11.84SGIRR44 pKa = 11.84KK45 pKa = 9.13SPYY48 pKa = 9.4LLKK51 pKa = 10.83SIIWKK56 pKa = 8.8EE57 pKa = 3.64QLRR60 pKa = 11.84QIEE63 pKa = 4.13QEE65 pKa = 3.88LAKK68 pKa = 10.24RR69 pKa = 11.84IHH71 pKa = 6.2NGRR74 pKa = 11.84RR75 pKa = 11.84YY76 pKa = 9.27PP77 pKa = 3.53
Molecular weight: 9.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
23668 |
37 |
3760 |
260.1 |
29.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.981 ± 0.368 | 0.722 ± 0.149 |
6.659 ± 0.134 | 7.318 ± 0.251 |
3.76 ± 0.175 | 7.318 ± 0.274 |
1.927 ± 0.263 | 5.564 ± 0.179 |
5.581 ± 0.315 | 7.952 ± 0.232 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.291 ± 0.151 | 4.935 ± 0.204 |
4.365 ± 0.17 | 4.441 ± 0.19 |
4.91 ± 0.258 | 5.607 ± 0.154 |
6.371 ± 0.23 | 6.591 ± 0.178 |
1.407 ± 0.132 | 3.3 ± 0.256 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
