
Peltaster fructicola
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetes incertae sedis; Peltaster
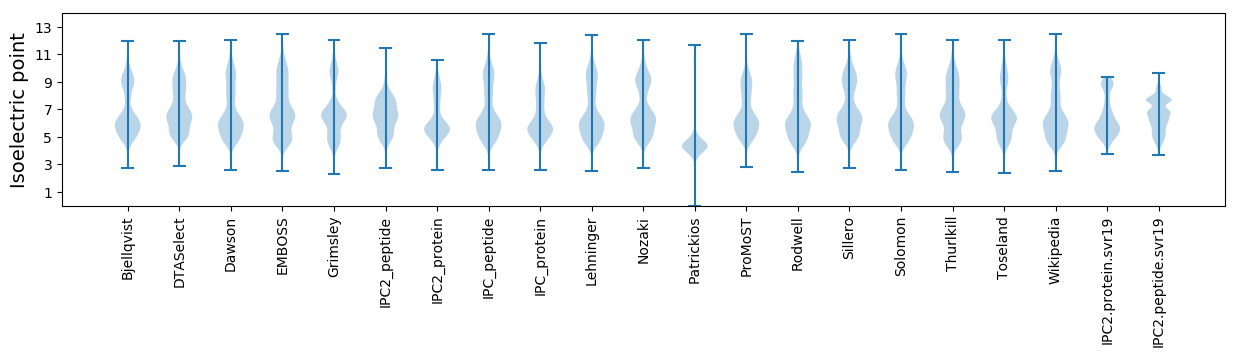
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
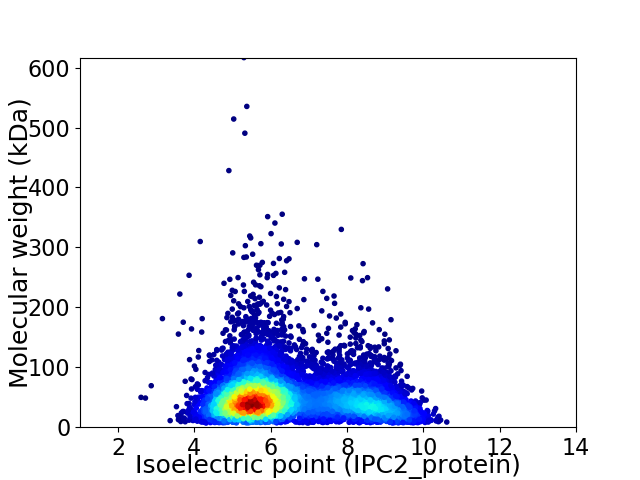
Virtual 2D-PAGE plot for 8072 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6H0XQ01|A0A6H0XQ01_9PEZI Phenylalanyl-tRNA synthetase OS=Peltaster fructicola OX=286661 GN=AMS68_002331 PE=3 SV=1
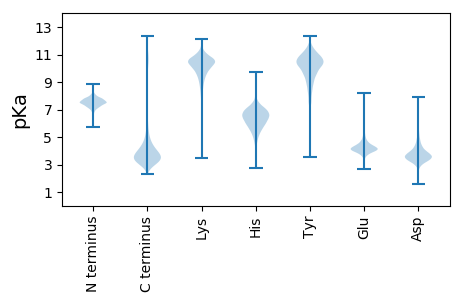
MM1 pKa = 7.8DD2 pKa = 5.45AEE4 pKa = 4.5NIYY7 pKa = 11.08DD8 pKa = 4.07EE9 pKa = 5.5IEE11 pKa = 4.47IEE13 pKa = 5.6DD14 pKa = 3.8MTFDD18 pKa = 3.93SALQMYY24 pKa = 8.48HH25 pKa = 5.92YY26 pKa = 8.49PCPCGDD32 pKa = 3.16RR33 pKa = 11.84FEE35 pKa = 5.85INIDD39 pKa = 3.58DD40 pKa = 4.31LRR42 pKa = 11.84DD43 pKa = 3.65GEE45 pKa = 4.73EE46 pKa = 3.84IAVCPSCSLMIRR58 pKa = 11.84VIFDD62 pKa = 3.16VDD64 pKa = 4.49DD65 pKa = 4.13LPKK68 pKa = 10.25PDD70 pKa = 5.18GGAQNAGQQAIATTVV85 pKa = 2.66
MM1 pKa = 7.8DD2 pKa = 5.45AEE4 pKa = 4.5NIYY7 pKa = 11.08DD8 pKa = 4.07EE9 pKa = 5.5IEE11 pKa = 4.47IEE13 pKa = 5.6DD14 pKa = 3.8MTFDD18 pKa = 3.93SALQMYY24 pKa = 8.48HH25 pKa = 5.92YY26 pKa = 8.49PCPCGDD32 pKa = 3.16RR33 pKa = 11.84FEE35 pKa = 5.85INIDD39 pKa = 3.58DD40 pKa = 4.31LRR42 pKa = 11.84DD43 pKa = 3.65GEE45 pKa = 4.73EE46 pKa = 3.84IAVCPSCSLMIRR58 pKa = 11.84VIFDD62 pKa = 3.16VDD64 pKa = 4.49DD65 pKa = 4.13LPKK68 pKa = 10.25PDD70 pKa = 5.18GGAQNAGQQAIATTVV85 pKa = 2.66
Molecular weight: 9.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6H0XSK2|A0A6H0XSK2_9PEZI C2H2-type domain-containing protein OS=Peltaster fructicola OX=286661 GN=AMS68_003231 PE=4 SV=1
MM1 pKa = 7.19FRR3 pKa = 11.84HH4 pKa = 5.99ALGISKK10 pKa = 10.17HH11 pKa = 5.43IALPDD16 pKa = 3.33VCFQCASIARR26 pKa = 11.84PIAQQRR32 pKa = 11.84SLQITARR39 pKa = 11.84RR40 pKa = 11.84QLASIRR46 pKa = 11.84GQALDD51 pKa = 3.18RR52 pKa = 11.84SYY54 pKa = 11.15FGSRR58 pKa = 11.84SPDD61 pKa = 2.9VSNAFRR67 pKa = 11.84HH68 pKa = 4.48TRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84KK75 pKa = 10.1DD76 pKa = 3.14EE77 pKa = 4.29EE78 pKa = 4.24KK79 pKa = 10.5PLPLDD84 pKa = 3.81ASSTLAQSASQAPARR99 pKa = 11.84SIRR102 pKa = 11.84RR103 pKa = 11.84LISTYY108 pKa = 10.79LALTKK113 pKa = 10.2PRR115 pKa = 11.84LAFLIVLTTTAAYY128 pKa = 10.1SIYY131 pKa = 9.81PVPAVLSSNVTAAPSLSTLTLAFLTTGTFLTIASANTLNMLFEE174 pKa = 4.73PAHH177 pKa = 5.9DD178 pKa = 3.73AKK180 pKa = 10.52MSRR183 pKa = 11.84TRR185 pKa = 11.84NRR187 pKa = 11.84PLVRR191 pKa = 11.84GLLSKK196 pKa = 10.35RR197 pKa = 11.84AAMVFAIATGVVGTGTLWLGVNPTTALLGAGNLILYY233 pKa = 9.24AFIYY237 pKa = 10.25TPFKK241 pKa = 10.48RR242 pKa = 11.84LHH244 pKa = 6.38PVNTWVGAIVGAIPPLMGWCAAGSQYY270 pKa = 8.43STKK273 pKa = 10.37AASPFDD279 pKa = 3.42IGTVWEE285 pKa = 4.11EE286 pKa = 4.66AKK288 pKa = 10.85DD289 pKa = 3.91LLCTEE294 pKa = 4.17QAIGGWLLAALLFAWQFPHH313 pKa = 7.14FFALSYY319 pKa = 10.83AVRR322 pKa = 11.84QEE324 pKa = 4.04YY325 pKa = 10.74AGAGYY330 pKa = 11.34KK331 pKa = 9.3MLTSTNVPMAARR343 pKa = 11.84VSFRR347 pKa = 11.84YY348 pKa = 9.48SLLMFPICFGLIVLQGDD365 pKa = 4.0RR366 pKa = 11.84ACFRR370 pKa = 11.84RR371 pKa = 11.84HH372 pKa = 5.16KK373 pKa = 10.25QCHH376 pKa = 3.82QCMDD380 pKa = 3.99AEE382 pKa = 4.08RR383 pKa = 11.84GMAILASQWRR393 pKa = 11.84EE394 pKa = 3.84RR395 pKa = 11.84KK396 pKa = 9.63CC397 pKa = 3.53
MM1 pKa = 7.19FRR3 pKa = 11.84HH4 pKa = 5.99ALGISKK10 pKa = 10.17HH11 pKa = 5.43IALPDD16 pKa = 3.33VCFQCASIARR26 pKa = 11.84PIAQQRR32 pKa = 11.84SLQITARR39 pKa = 11.84RR40 pKa = 11.84QLASIRR46 pKa = 11.84GQALDD51 pKa = 3.18RR52 pKa = 11.84SYY54 pKa = 11.15FGSRR58 pKa = 11.84SPDD61 pKa = 2.9VSNAFRR67 pKa = 11.84HH68 pKa = 4.48TRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84KK75 pKa = 10.1DD76 pKa = 3.14EE77 pKa = 4.29EE78 pKa = 4.24KK79 pKa = 10.5PLPLDD84 pKa = 3.81ASSTLAQSASQAPARR99 pKa = 11.84SIRR102 pKa = 11.84RR103 pKa = 11.84LISTYY108 pKa = 10.79LALTKK113 pKa = 10.2PRR115 pKa = 11.84LAFLIVLTTTAAYY128 pKa = 10.1SIYY131 pKa = 9.81PVPAVLSSNVTAAPSLSTLTLAFLTTGTFLTIASANTLNMLFEE174 pKa = 4.73PAHH177 pKa = 5.9DD178 pKa = 3.73AKK180 pKa = 10.52MSRR183 pKa = 11.84TRR185 pKa = 11.84NRR187 pKa = 11.84PLVRR191 pKa = 11.84GLLSKK196 pKa = 10.35RR197 pKa = 11.84AAMVFAIATGVVGTGTLWLGVNPTTALLGAGNLILYY233 pKa = 9.24AFIYY237 pKa = 10.25TPFKK241 pKa = 10.48RR242 pKa = 11.84LHH244 pKa = 6.38PVNTWVGAIVGAIPPLMGWCAAGSQYY270 pKa = 8.43STKK273 pKa = 10.37AASPFDD279 pKa = 3.42IGTVWEE285 pKa = 4.11EE286 pKa = 4.66AKK288 pKa = 10.85DD289 pKa = 3.91LLCTEE294 pKa = 4.17QAIGGWLLAALLFAWQFPHH313 pKa = 7.14FFALSYY319 pKa = 10.83AVRR322 pKa = 11.84QEE324 pKa = 4.04YY325 pKa = 10.74AGAGYY330 pKa = 11.34KK331 pKa = 9.3MLTSTNVPMAARR343 pKa = 11.84VSFRR347 pKa = 11.84YY348 pKa = 9.48SLLMFPICFGLIVLQGDD365 pKa = 4.0RR366 pKa = 11.84ACFRR370 pKa = 11.84RR371 pKa = 11.84HH372 pKa = 5.16KK373 pKa = 10.25QCHH376 pKa = 3.82QCMDD380 pKa = 3.99AEE382 pKa = 4.08RR383 pKa = 11.84GMAILASQWRR393 pKa = 11.84EE394 pKa = 3.84RR395 pKa = 11.84KK396 pKa = 9.63CC397 pKa = 3.53
Molecular weight: 43.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4031696 |
66 |
5569 |
499.5 |
55.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.286 ± 0.023 | 1.127 ± 0.011 |
5.969 ± 0.021 | 5.995 ± 0.026 |
3.46 ± 0.016 | 6.459 ± 0.025 |
2.469 ± 0.012 | 4.86 ± 0.018 |
4.83 ± 0.023 | 8.889 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.151 ± 0.01 | 3.545 ± 0.014 |
5.611 ± 0.026 | 4.467 ± 0.021 |
6.126 ± 0.03 | 8.163 ± 0.034 |
6.367 ± 0.025 | 6.097 ± 0.021 |
1.372 ± 0.01 | 2.759 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
