
Caulobacter phage CcrColossus
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Dolichocephalovirinae; Colossusvirus; Caulobacter virus CcrColossus
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
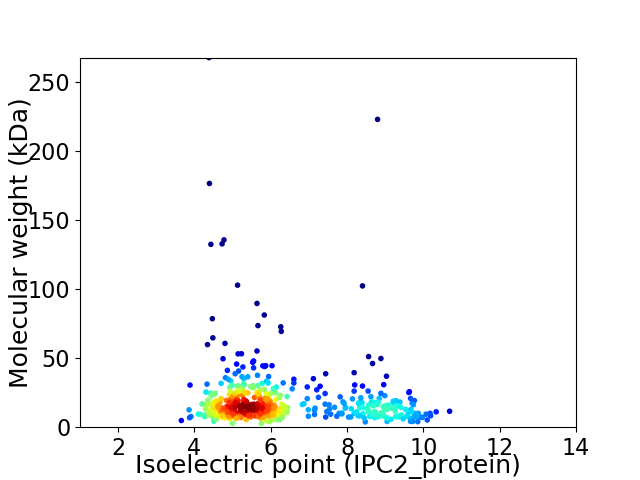
Virtual 2D-PAGE plot for 448 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
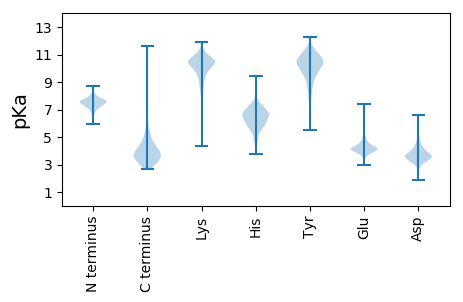
Protein with the lowest isoelectric point:
>tr|K4JRI5|K4JRI5_9CAUD Uncharacterized protein OS=Caulobacter phage CcrColossus OX=1211640 GN=CcrColossus_gp044 PE=4 SV=1
MM1 pKa = 7.6TSQLCIADD9 pKa = 3.78EE10 pKa = 4.02NAALDD15 pKa = 3.87FLARR19 pKa = 11.84AVPALCDD26 pKa = 3.76AGIMPPTLDD35 pKa = 4.18ILDD38 pKa = 4.15SLAANPNAYY47 pKa = 9.84GLSRR51 pKa = 11.84VPTLAPLAAEE61 pKa = 4.01WAGPYY66 pKa = 7.66QTQANDD72 pKa = 3.89GEE74 pKa = 4.91GPTTVRR80 pKa = 11.84ALIVAALGADD90 pKa = 3.32EE91 pKa = 5.49SGNFDD96 pKa = 3.21IGFRR100 pKa = 11.84TYY102 pKa = 11.17AVNDD106 pKa = 3.81TEE108 pKa = 4.23HH109 pKa = 6.89WNLAGTPLEE118 pKa = 4.28GVPAYY123 pKa = 10.35SLYY126 pKa = 10.76LIRR129 pKa = 11.84EE130 pKa = 4.49GEE132 pKa = 4.03QTYY135 pKa = 8.51MCSLTPSSYY144 pKa = 11.24AYY146 pKa = 10.25GIQNVFLYY154 pKa = 9.73PDD156 pKa = 3.62QEE158 pKa = 4.23GLPAVNEE165 pKa = 4.13YY166 pKa = 10.83GQTAVEE172 pKa = 4.48AYY174 pKa = 9.22MDD176 pKa = 4.15AEE178 pKa = 4.15NPEE181 pKa = 4.19YY182 pKa = 10.56EE183 pKa = 4.42AVEE186 pKa = 4.08DD187 pKa = 4.37TYY189 pKa = 11.16FGYY192 pKa = 9.98MGDD195 pKa = 3.93RR196 pKa = 11.84EE197 pKa = 4.21ALDD200 pKa = 3.19RR201 pKa = 11.84RR202 pKa = 11.84EE203 pKa = 4.21AEE205 pKa = 4.06EE206 pKa = 5.01GDD208 pKa = 3.46VSEE211 pKa = 5.18RR212 pKa = 11.84LSFDD216 pKa = 2.83KK217 pKa = 11.15SVFRR221 pKa = 11.84VDD223 pKa = 5.33LSTLDD228 pKa = 3.73GFAASPANPRR238 pKa = 11.84EE239 pKa = 4.02TTAFADD245 pKa = 4.2FEE247 pKa = 4.46TLAAAFYY254 pKa = 10.55KK255 pKa = 10.44AACDD259 pKa = 4.6SAWEE263 pKa = 4.0DD264 pKa = 3.05AKK266 pKa = 11.28EE267 pKa = 3.98YY268 pKa = 10.95VSGNATEE275 pKa = 4.06PRR277 pKa = 11.84ILTGCC282 pKa = 3.89
MM1 pKa = 7.6TSQLCIADD9 pKa = 3.78EE10 pKa = 4.02NAALDD15 pKa = 3.87FLARR19 pKa = 11.84AVPALCDD26 pKa = 3.76AGIMPPTLDD35 pKa = 4.18ILDD38 pKa = 4.15SLAANPNAYY47 pKa = 9.84GLSRR51 pKa = 11.84VPTLAPLAAEE61 pKa = 4.01WAGPYY66 pKa = 7.66QTQANDD72 pKa = 3.89GEE74 pKa = 4.91GPTTVRR80 pKa = 11.84ALIVAALGADD90 pKa = 3.32EE91 pKa = 5.49SGNFDD96 pKa = 3.21IGFRR100 pKa = 11.84TYY102 pKa = 11.17AVNDD106 pKa = 3.81TEE108 pKa = 4.23HH109 pKa = 6.89WNLAGTPLEE118 pKa = 4.28GVPAYY123 pKa = 10.35SLYY126 pKa = 10.76LIRR129 pKa = 11.84EE130 pKa = 4.49GEE132 pKa = 4.03QTYY135 pKa = 8.51MCSLTPSSYY144 pKa = 11.24AYY146 pKa = 10.25GIQNVFLYY154 pKa = 9.73PDD156 pKa = 3.62QEE158 pKa = 4.23GLPAVNEE165 pKa = 4.13YY166 pKa = 10.83GQTAVEE172 pKa = 4.48AYY174 pKa = 9.22MDD176 pKa = 4.15AEE178 pKa = 4.15NPEE181 pKa = 4.19YY182 pKa = 10.56EE183 pKa = 4.42AVEE186 pKa = 4.08DD187 pKa = 4.37TYY189 pKa = 11.16FGYY192 pKa = 9.98MGDD195 pKa = 3.93RR196 pKa = 11.84EE197 pKa = 4.21ALDD200 pKa = 3.19RR201 pKa = 11.84RR202 pKa = 11.84EE203 pKa = 4.21AEE205 pKa = 4.06EE206 pKa = 5.01GDD208 pKa = 3.46VSEE211 pKa = 5.18RR212 pKa = 11.84LSFDD216 pKa = 2.83KK217 pKa = 11.15SVFRR221 pKa = 11.84VDD223 pKa = 5.33LSTLDD228 pKa = 3.73GFAASPANPRR238 pKa = 11.84EE239 pKa = 4.02TTAFADD245 pKa = 4.2FEE247 pKa = 4.46TLAAAFYY254 pKa = 10.55KK255 pKa = 10.44AACDD259 pKa = 4.6SAWEE263 pKa = 4.0DD264 pKa = 3.05AKK266 pKa = 11.28EE267 pKa = 3.98YY268 pKa = 10.95VSGNATEE275 pKa = 4.06PRR277 pKa = 11.84ILTGCC282 pKa = 3.89
Molecular weight: 30.48 kDa
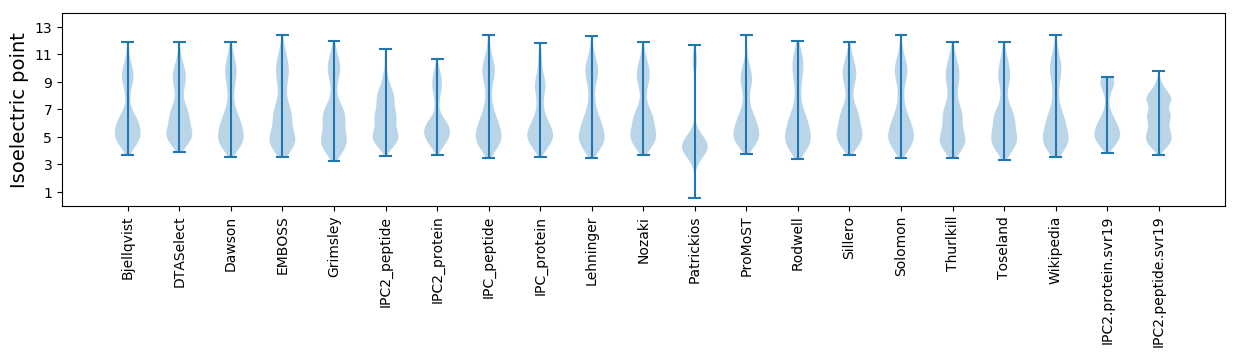
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4JUG3|K4JUG3_9CAUD Uncharacterized protein OS=Caulobacter phage CcrColossus OX=1211640 GN=CcrColossus_gp105 PE=4 SV=1
MM1 pKa = 7.23PTRR4 pKa = 11.84ARR6 pKa = 11.84RR7 pKa = 11.84SRR9 pKa = 11.84AKK11 pKa = 10.19RR12 pKa = 11.84ALAPGQQEE20 pKa = 4.36KK21 pKa = 11.01ALGASPGIYY30 pKa = 9.13IVMEE34 pKa = 4.68LGMHH38 pKa = 7.2PYY40 pKa = 10.35SPPTDD45 pKa = 3.01IGRR48 pKa = 11.84TIAGHH53 pKa = 5.3EE54 pKa = 4.24VHH56 pKa = 6.93HH57 pKa = 6.25LTADD61 pKa = 3.65CPRR64 pKa = 11.84NHH66 pKa = 6.33SRR68 pKa = 11.84RR69 pKa = 11.84AAKK72 pKa = 10.2RR73 pKa = 11.84LRR75 pKa = 11.84HH76 pKa = 6.24AARR79 pKa = 11.84QYY81 pKa = 11.15DD82 pKa = 3.68RR83 pKa = 11.84QLICQEE89 pKa = 3.89VANFLDD95 pKa = 3.83ASLAAA100 pKa = 4.97
MM1 pKa = 7.23PTRR4 pKa = 11.84ARR6 pKa = 11.84RR7 pKa = 11.84SRR9 pKa = 11.84AKK11 pKa = 10.19RR12 pKa = 11.84ALAPGQQEE20 pKa = 4.36KK21 pKa = 11.01ALGASPGIYY30 pKa = 9.13IVMEE34 pKa = 4.68LGMHH38 pKa = 7.2PYY40 pKa = 10.35SPPTDD45 pKa = 3.01IGRR48 pKa = 11.84TIAGHH53 pKa = 5.3EE54 pKa = 4.24VHH56 pKa = 6.93HH57 pKa = 6.25LTADD61 pKa = 3.65CPRR64 pKa = 11.84NHH66 pKa = 6.33SRR68 pKa = 11.84RR69 pKa = 11.84AAKK72 pKa = 10.2RR73 pKa = 11.84LRR75 pKa = 11.84HH76 pKa = 6.24AARR79 pKa = 11.84QYY81 pKa = 11.15DD82 pKa = 3.68RR83 pKa = 11.84QLICQEE89 pKa = 3.89VANFLDD95 pKa = 3.83ASLAAA100 pKa = 4.97
Molecular weight: 11.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
83195 |
22 |
2750 |
185.7 |
20.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.789 ± 0.206 | 0.95 ± 0.058 |
6.433 ± 0.152 | 6.294 ± 0.203 |
3.761 ± 0.079 | 8.176 ± 0.448 |
2.263 ± 0.1 | 4.956 ± 0.074 |
4.487 ± 0.156 | 7.998 ± 0.142 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.483 ± 0.077 | 3.231 ± 0.109 |
5.212 ± 0.103 | 3.434 ± 0.084 |
6.69 ± 0.235 | 5.369 ± 0.142 |
5.892 ± 0.194 | 6.69 ± 0.115 |
1.688 ± 0.064 | 3.203 ± 0.089 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
