
Loktanella sp. 3ANDIMAR09
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Loktanella; unclassified Loktanella
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
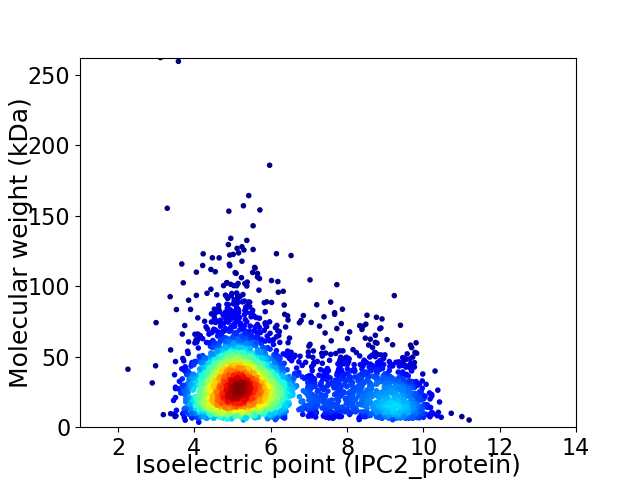
Virtual 2D-PAGE plot for 3527 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
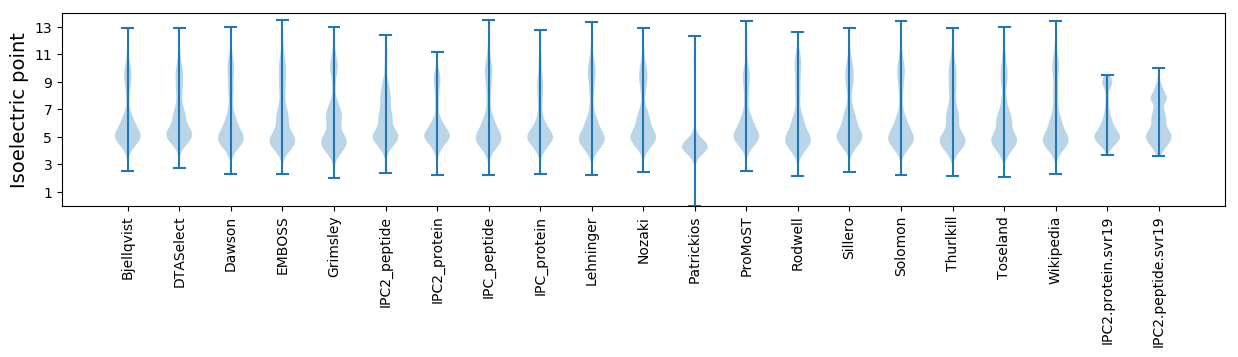
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q2YX32|A0A0Q2YX32_9RHOB Uncharacterized protein OS=Loktanella sp. 3ANDIMAR09 OX=1225657 GN=AN189_14305 PE=4 SV=1
MM1 pKa = 7.62EE2 pKa = 5.32KK3 pKa = 10.57FRR5 pKa = 11.84WTLSARR11 pKa = 11.84ASRR14 pKa = 11.84AAAFLAFLLQICVATVAGATPFTMTVPGTGVQLPAEE50 pKa = 4.34YY51 pKa = 9.08PQAGGVAIVMTGVNGNIYY69 pKa = 10.12YY70 pKa = 10.25QFSNPDD76 pKa = 3.04GAFVGFQNTGTPTRR90 pKa = 11.84FRR92 pKa = 11.84GNPFTINDD100 pKa = 5.89PIPLDD105 pKa = 3.93CGFRR109 pKa = 11.84SCTDD113 pKa = 3.26YY114 pKa = 11.34FGGAIARR121 pKa = 11.84VDD123 pKa = 3.32IRR125 pKa = 11.84FSALDD130 pKa = 3.47GDD132 pKa = 4.42TQVGGFDD139 pKa = 3.81RR140 pKa = 11.84NDD142 pKa = 2.76IFLILNGVNVGNWSDD157 pKa = 3.53VTTQATNEE165 pKa = 3.99AGTTSFSTQQGFGNNTFDD183 pKa = 3.75TGWFSSTNAALLGNILQANQTVTQVFDD210 pKa = 4.37RR211 pKa = 11.84DD212 pKa = 3.84PNDD215 pKa = 3.49NFWDD219 pKa = 4.62FTRR222 pKa = 11.84GNSLPQDD229 pKa = 4.94DD230 pKa = 5.0IRR232 pKa = 11.84TIAPGYY238 pKa = 8.98EE239 pKa = 3.84FEE241 pKa = 5.79KK242 pKa = 10.61IIDD245 pKa = 4.1SADD248 pKa = 3.1LTYY251 pKa = 11.15AQVGDD256 pKa = 4.34IIDD259 pKa = 3.34YY260 pKa = 10.62SYY262 pKa = 11.51VVRR265 pKa = 11.84NIGSVDD271 pKa = 3.0IDD273 pKa = 5.08DD274 pKa = 4.06ITVEE278 pKa = 4.64DD279 pKa = 4.06DD280 pKa = 4.15RR281 pKa = 11.84IPNVVCPAPPQNSLRR296 pKa = 11.84RR297 pKa = 11.84TRR299 pKa = 11.84GGDD302 pKa = 3.11PTANEE307 pKa = 4.18LTCTGTYY314 pKa = 10.18RR315 pKa = 11.84ITQADD320 pKa = 3.3IDD322 pKa = 4.18AGQVTNIASANGTPEE337 pKa = 3.82FGEE340 pKa = 4.3LGEE343 pKa = 4.41VTDD346 pKa = 4.15TATTTGPALVNTITLDD362 pKa = 3.36KK363 pKa = 11.05VGAPDD368 pKa = 3.52PFGPVGSQVTYY379 pKa = 10.45TLTATNTGTSTLRR392 pKa = 11.84DD393 pKa = 3.68VLITDD398 pKa = 4.08PRR400 pKa = 11.84TPGQTCTFPTLLPLSADD417 pKa = 3.31NATNTASCTVTYY429 pKa = 9.69TVTQADD435 pKa = 3.29VDD437 pKa = 3.99AFALNGTTLDD447 pKa = 3.25NTATVRR453 pKa = 11.84ATPPNGPAITATASDD468 pKa = 4.55ALPGQPAVVSLTLVKK483 pKa = 10.46SALQSTYY490 pKa = 11.17DD491 pKa = 3.41AVGDD495 pKa = 3.91VIDD498 pKa = 4.48YY499 pKa = 10.73RR500 pKa = 11.84FTINNTGTVTWPGPPTLTDD519 pKa = 3.49TLTGGASCPAGPVAPGGTVVCEE541 pKa = 3.81ASYY544 pKa = 10.31TILQADD550 pKa = 4.48LDD552 pKa = 4.17AGVRR556 pKa = 11.84PNEE559 pKa = 3.63ATAAITVAGQTAQATDD575 pKa = 4.05DD576 pKa = 3.83VTVTAVVTTGLTLSKK591 pKa = 10.33ALQAGSPNTFDD602 pKa = 3.44APGVVLTYY610 pKa = 11.25NFVLTNTGTVTLTNPVVDD628 pKa = 5.05DD629 pKa = 3.73ATTAPTVTPVPVTCPAGPIAPGGTLTCSADD659 pKa = 3.69YY660 pKa = 11.1AVTQADD666 pKa = 4.01LDD668 pKa = 4.09AGAVSNTATAAATAPPGAGGGQVLSGPQSLTVTAEE703 pKa = 3.53RR704 pKa = 11.84RR705 pKa = 11.84PALNIVKK712 pKa = 8.15TAPTLTALQFSALPVVTYY730 pKa = 9.53TYY732 pKa = 11.36DD733 pKa = 3.26VTNSGNVTIPGPITVTDD750 pKa = 4.6DD751 pKa = 3.43KK752 pKa = 11.39LSPQTFTCTGAAIAVGDD769 pKa = 4.35SVQCTATYY777 pKa = 10.35QVTPTDD783 pKa = 3.43VARR786 pKa = 11.84QSVRR790 pKa = 11.84NTAFATGGDD799 pKa = 3.77GTVSNTDD806 pKa = 3.5DD807 pKa = 4.63AVIAPAGRR815 pKa = 11.84LDD817 pKa = 3.5IQLEE821 pKa = 4.29KK822 pKa = 10.79QAVTTDD828 pKa = 4.01FAAVGDD834 pKa = 4.22PVDD837 pKa = 4.0FTLTFTNTGDD847 pKa = 3.36FTLFPEE853 pKa = 4.67TDD855 pKa = 2.97SSALDD860 pKa = 3.51RR861 pKa = 11.84YY862 pKa = 10.18EE863 pKa = 3.86LTDD866 pKa = 4.05PGVSGLDD873 pKa = 3.16SCIIPNPLFPQGGAQTPTQFTCTVTRR899 pKa = 11.84IITQADD905 pKa = 3.34VDD907 pKa = 3.95AGSYY911 pKa = 10.12TNTAFLTMIYY921 pKa = 10.42NQGQVNEE928 pKa = 4.33EE929 pKa = 4.32TQVVPPASDD938 pKa = 3.54TVPLNANVVPSFTLTKK954 pKa = 9.97TATNDD959 pKa = 3.13YY960 pKa = 10.06GAVGDD965 pKa = 4.06TINYY969 pKa = 9.43DD970 pKa = 3.73FEE972 pKa = 4.53VTNTSVLTLTNVVLTDD988 pKa = 3.76PKK990 pKa = 10.24IGSISCVGSATANEE1004 pKa = 4.5TGTIAPGASALCTGSYY1020 pKa = 10.42EE1021 pKa = 4.17VTQADD1026 pKa = 3.85LDD1028 pKa = 4.0AEE1030 pKa = 4.38QVVNTVSALAQTQAAGSVTQTAQEE1054 pKa = 3.93ITPIDD1059 pKa = 3.56PAAATRR1065 pKa = 11.84ALSLTKK1071 pKa = 9.94IVTVRR1076 pKa = 11.84EE1077 pKa = 4.05TGDD1080 pKa = 3.25ATFTAPGQTADD1091 pKa = 3.42YY1092 pKa = 10.46SFVVEE1097 pKa = 4.05NTGNLTLDD1105 pKa = 3.98NIAVVDD1111 pKa = 3.88ADD1113 pKa = 5.17LGYY1116 pKa = 8.65TCQIPTLAPGATDD1129 pKa = 3.28TATCSASRR1137 pKa = 11.84VLTQADD1143 pKa = 4.05FDD1145 pKa = 4.19AGSYY1149 pKa = 10.38QNDD1152 pKa = 3.21AVARR1156 pKa = 11.84AAGADD1161 pKa = 3.9DD1162 pKa = 4.66APASATATAQGRR1174 pKa = 11.84IASFDD1179 pKa = 3.84FVKK1182 pKa = 10.61SAPASFAVAGEE1193 pKa = 4.11VVDD1196 pKa = 5.73FVFAFTNTGTVTLTDD1211 pKa = 3.33ITIVDD1216 pKa = 4.02PLLGVAYY1223 pKa = 9.72ACEE1226 pKa = 4.14ITSIAPGVTDD1236 pKa = 3.68NSCFGSYY1243 pKa = 8.4EE1244 pKa = 4.02VQPADD1249 pKa = 3.25VDD1251 pKa = 3.56AGQIQNDD1258 pKa = 3.37ASFTGVGLDD1267 pKa = 3.62GAPISGDD1274 pKa = 3.1DD1275 pKa = 3.71TVIVTGPAEE1284 pKa = 4.29APALTVTKK1292 pKa = 10.74VEE1294 pKa = 4.53DD1295 pKa = 4.42DD1296 pKa = 3.67GTGTFDD1302 pKa = 4.09NLPATEE1308 pKa = 4.52SYY1310 pKa = 11.29SFVVTNTGNVTLTDD1324 pKa = 4.33LSLVDD1329 pKa = 4.6DD1330 pKa = 4.37LTGFSCALPDD1340 pKa = 4.23LAAGAATSLCADD1352 pKa = 4.69GAPLTDD1358 pKa = 4.1SYY1360 pKa = 11.76TVLQTDD1366 pKa = 3.33IDD1368 pKa = 4.06AGALEE1373 pKa = 4.29NTATATGRR1381 pKa = 11.84TTRR1384 pKa = 11.84GAGVPVEE1391 pKa = 4.64ASDD1394 pKa = 3.75TVTLQGPAQLPVISMVKK1411 pKa = 8.9TATAGAGFSALGDD1424 pKa = 3.88TVSYY1428 pKa = 10.78DD1429 pKa = 3.49YY1430 pKa = 11.34TVTNDD1435 pKa = 3.27GNITLTAPITVTDD1448 pKa = 3.63DD1449 pKa = 3.36RR1450 pKa = 11.84VTVSCPALPAAGLAPGADD1468 pKa = 3.51IVCTATDD1475 pKa = 3.61TVDD1478 pKa = 4.66QADD1481 pKa = 4.04LDD1483 pKa = 3.85AGEE1486 pKa = 4.61IVNTATANVRR1496 pKa = 11.84QPVSPSATYY1505 pKa = 9.4PGGVAEE1511 pKa = 4.32VSSNTEE1517 pKa = 4.15TVTVTADD1524 pKa = 3.37QQPALAITKK1533 pKa = 9.63RR1534 pKa = 11.84VKK1536 pKa = 10.48PGTATSFDD1544 pKa = 3.83APGDD1548 pKa = 3.64LSDD1551 pKa = 5.02PGNPNNLVYY1560 pKa = 10.65EE1561 pKa = 4.85FIVEE1565 pKa = 3.98NTGNVTTTDD1574 pKa = 5.12PITVTDD1580 pKa = 4.45PLISATPLVCTTDD1593 pKa = 3.58PLAPGGTVTCEE1604 pKa = 3.57LPYY1607 pKa = 10.99APTQEE1612 pKa = 4.23DD1613 pKa = 3.26VDD1615 pKa = 3.97RR1616 pKa = 11.84GFFANSADD1624 pKa = 3.17AATRR1628 pKa = 11.84FNGAPVQTPAPGTEE1642 pKa = 4.08TVNAVQRR1649 pKa = 11.84PALTIVKK1656 pKa = 8.64TLEE1659 pKa = 4.05DD1660 pKa = 3.77LAIFSPGEE1668 pKa = 3.38IATYY1672 pKa = 10.34RR1673 pKa = 11.84FYY1675 pKa = 10.89VTNTGNTTITGPVTVTDD1692 pKa = 3.95NKK1694 pKa = 10.1IAGPIDD1700 pKa = 4.37CGPGDD1705 pKa = 4.53LAPGDD1710 pKa = 4.05STACLATYY1718 pKa = 8.42TVTVNDD1724 pKa = 3.58VNAGSVTNSASASNGVTTSDD1744 pKa = 3.95PQSLTIPVGAEE1755 pKa = 3.6PTLSIVKK1762 pKa = 7.61TAPGGVFAQAGDD1774 pKa = 3.71VIAYY1778 pKa = 7.63EE1779 pKa = 4.19FLVTNTSSGPIPPAFVQPVTVIDD1802 pKa = 4.5DD1803 pKa = 4.56KK1804 pKa = 10.78ITTPIVCPVVDD1815 pKa = 4.53GANPLDD1821 pKa = 4.3PGEE1824 pKa = 5.37SITCTGSYY1832 pKa = 8.94TVQQADD1838 pKa = 3.71LDD1840 pKa = 4.0AVRR1843 pKa = 11.84PGFTRR1848 pKa = 11.84GFVINNAFARR1858 pKa = 11.84SEE1860 pKa = 4.04FAGEE1864 pKa = 4.09LVEE1867 pKa = 5.17SPVDD1871 pKa = 3.64QVIITADD1878 pKa = 3.75SVPLLGVVKK1887 pKa = 9.3TAEE1890 pKa = 4.09NQTNPGQDD1898 pKa = 2.86AAAGDD1903 pKa = 4.08VVAFTIVSQNDD1914 pKa = 4.02GPQTLTDD1921 pKa = 3.57VRR1923 pKa = 11.84VTDD1926 pKa = 4.13PFLGALSCDD1935 pKa = 2.94IASPVTLAPGAALTCTGSYY1954 pKa = 9.35TVTQADD1960 pKa = 3.47VDD1962 pKa = 3.87AGGPISNTADD1972 pKa = 3.15AAATTTQGEE1981 pKa = 4.57AVSGTDD1987 pKa = 2.95TTTYY1991 pKa = 10.21PVAAPLPAMQVAKK2004 pKa = 10.5ALEE2007 pKa = 4.6PGSGQNDD2014 pKa = 4.23YY2015 pKa = 9.48NTVGEE2020 pKa = 4.4DD2021 pKa = 3.0LTFRR2025 pKa = 11.84VTVTNTGNITLNDD2038 pKa = 3.49VAVTDD2043 pKa = 4.26DD2044 pKa = 4.99LVPGTCTVPTLAPGASDD2061 pKa = 3.66TSCVFVYY2068 pKa = 9.15TVQQADD2074 pKa = 3.43IDD2076 pKa = 4.05RR2077 pKa = 11.84GTITNIATAQAQPANPGAPEE2097 pKa = 4.85LIEE2100 pKa = 4.45TGQSDD2105 pKa = 3.56ITGPAAEE2112 pKa = 4.87PGVGLSKK2119 pKa = 9.84TGSGEE2124 pKa = 3.92VTASDD2129 pKa = 4.24GSPTFNLPNQSLTYY2143 pKa = 9.93VYY2145 pKa = 10.68EE2146 pKa = 4.11VVNLGNVTLNQLPAITDD2163 pKa = 3.47DD2164 pKa = 4.96RR2165 pKa = 11.84IDD2167 pKa = 4.58SVTCDD2172 pKa = 3.94PLPAAGLLPGEE2183 pKa = 4.32VLICRR2188 pKa = 11.84GSDD2191 pKa = 2.95TVDD2194 pKa = 3.31QNDD2197 pKa = 2.94VDD2199 pKa = 4.46AGFVTNIADD2208 pKa = 3.46VTVANDD2214 pKa = 3.35YY2215 pKa = 11.34GGAPLTAQAQEE2226 pKa = 4.5TITATVAPDD2235 pKa = 3.41LAITKK2240 pKa = 8.44TPSITQNATAGTEE2253 pKa = 3.79VTYY2256 pKa = 10.26SYY2258 pKa = 10.97RR2259 pKa = 11.84VQNPGNVTLTDD2270 pKa = 3.05ITLTDD2275 pKa = 4.01FHH2277 pKa = 6.72TSAAGVVDD2285 pKa = 5.42LPISDD2290 pKa = 4.21GGVIATLAPGEE2301 pKa = 4.35EE2302 pKa = 4.26ATLTATYY2309 pKa = 9.51TVTQDD2314 pKa = 5.55DD2315 pKa = 4.23IDD2317 pKa = 3.83SGNAVRR2323 pKa = 11.84NTVRR2327 pKa = 11.84VTSTLPGGDD2336 pKa = 4.07AGPAPLTRR2344 pKa = 11.84ARR2346 pKa = 11.84VDD2348 pKa = 4.2LDD2350 pKa = 3.38PQRR2353 pKa = 11.84PAMEE2357 pKa = 5.12AIKK2360 pKa = 9.0TIPSSPVPPVPGSQITYY2377 pKa = 10.47QITLTNTGNVTLGDD2391 pKa = 3.9PVLTDD2396 pKa = 3.08TVRR2399 pKa = 11.84RR2400 pKa = 11.84INTDD2404 pKa = 2.94ALAGEE2409 pKa = 4.37PAVALTGGDD2418 pKa = 3.2TGTPGLFEE2426 pKa = 4.23VGEE2429 pKa = 4.16VRR2431 pKa = 11.84TYY2433 pKa = 10.99AVSYY2437 pKa = 9.64TLTQDD2442 pKa = 5.01DD2443 pKa = 3.79IDD2445 pKa = 5.23AGGVANSVTATADD2458 pKa = 3.52DD2459 pKa = 4.26PLGNPVTEE2467 pKa = 4.32VSDD2470 pKa = 3.63NGAGDD2475 pKa = 4.03GDD2477 pKa = 4.56DD2478 pKa = 3.43PTLFVIAPTPALEE2491 pKa = 4.23VIKK2494 pKa = 10.81VISQPASAVGEE2505 pKa = 4.2TVVFEE2510 pKa = 3.96ITATNTGNVTLLDD2523 pKa = 3.83AALTDD2528 pKa = 3.37SFTRR2532 pKa = 11.84LDD2534 pKa = 3.72GTPIPATPP2542 pKa = 3.65
MM1 pKa = 7.62EE2 pKa = 5.32KK3 pKa = 10.57FRR5 pKa = 11.84WTLSARR11 pKa = 11.84ASRR14 pKa = 11.84AAAFLAFLLQICVATVAGATPFTMTVPGTGVQLPAEE50 pKa = 4.34YY51 pKa = 9.08PQAGGVAIVMTGVNGNIYY69 pKa = 10.12YY70 pKa = 10.25QFSNPDD76 pKa = 3.04GAFVGFQNTGTPTRR90 pKa = 11.84FRR92 pKa = 11.84GNPFTINDD100 pKa = 5.89PIPLDD105 pKa = 3.93CGFRR109 pKa = 11.84SCTDD113 pKa = 3.26YY114 pKa = 11.34FGGAIARR121 pKa = 11.84VDD123 pKa = 3.32IRR125 pKa = 11.84FSALDD130 pKa = 3.47GDD132 pKa = 4.42TQVGGFDD139 pKa = 3.81RR140 pKa = 11.84NDD142 pKa = 2.76IFLILNGVNVGNWSDD157 pKa = 3.53VTTQATNEE165 pKa = 3.99AGTTSFSTQQGFGNNTFDD183 pKa = 3.75TGWFSSTNAALLGNILQANQTVTQVFDD210 pKa = 4.37RR211 pKa = 11.84DD212 pKa = 3.84PNDD215 pKa = 3.49NFWDD219 pKa = 4.62FTRR222 pKa = 11.84GNSLPQDD229 pKa = 4.94DD230 pKa = 5.0IRR232 pKa = 11.84TIAPGYY238 pKa = 8.98EE239 pKa = 3.84FEE241 pKa = 5.79KK242 pKa = 10.61IIDD245 pKa = 4.1SADD248 pKa = 3.1LTYY251 pKa = 11.15AQVGDD256 pKa = 4.34IIDD259 pKa = 3.34YY260 pKa = 10.62SYY262 pKa = 11.51VVRR265 pKa = 11.84NIGSVDD271 pKa = 3.0IDD273 pKa = 5.08DD274 pKa = 4.06ITVEE278 pKa = 4.64DD279 pKa = 4.06DD280 pKa = 4.15RR281 pKa = 11.84IPNVVCPAPPQNSLRR296 pKa = 11.84RR297 pKa = 11.84TRR299 pKa = 11.84GGDD302 pKa = 3.11PTANEE307 pKa = 4.18LTCTGTYY314 pKa = 10.18RR315 pKa = 11.84ITQADD320 pKa = 3.3IDD322 pKa = 4.18AGQVTNIASANGTPEE337 pKa = 3.82FGEE340 pKa = 4.3LGEE343 pKa = 4.41VTDD346 pKa = 4.15TATTTGPALVNTITLDD362 pKa = 3.36KK363 pKa = 11.05VGAPDD368 pKa = 3.52PFGPVGSQVTYY379 pKa = 10.45TLTATNTGTSTLRR392 pKa = 11.84DD393 pKa = 3.68VLITDD398 pKa = 4.08PRR400 pKa = 11.84TPGQTCTFPTLLPLSADD417 pKa = 3.31NATNTASCTVTYY429 pKa = 9.69TVTQADD435 pKa = 3.29VDD437 pKa = 3.99AFALNGTTLDD447 pKa = 3.25NTATVRR453 pKa = 11.84ATPPNGPAITATASDD468 pKa = 4.55ALPGQPAVVSLTLVKK483 pKa = 10.46SALQSTYY490 pKa = 11.17DD491 pKa = 3.41AVGDD495 pKa = 3.91VIDD498 pKa = 4.48YY499 pKa = 10.73RR500 pKa = 11.84FTINNTGTVTWPGPPTLTDD519 pKa = 3.49TLTGGASCPAGPVAPGGTVVCEE541 pKa = 3.81ASYY544 pKa = 10.31TILQADD550 pKa = 4.48LDD552 pKa = 4.17AGVRR556 pKa = 11.84PNEE559 pKa = 3.63ATAAITVAGQTAQATDD575 pKa = 4.05DD576 pKa = 3.83VTVTAVVTTGLTLSKK591 pKa = 10.33ALQAGSPNTFDD602 pKa = 3.44APGVVLTYY610 pKa = 11.25NFVLTNTGTVTLTNPVVDD628 pKa = 5.05DD629 pKa = 3.73ATTAPTVTPVPVTCPAGPIAPGGTLTCSADD659 pKa = 3.69YY660 pKa = 11.1AVTQADD666 pKa = 4.01LDD668 pKa = 4.09AGAVSNTATAAATAPPGAGGGQVLSGPQSLTVTAEE703 pKa = 3.53RR704 pKa = 11.84RR705 pKa = 11.84PALNIVKK712 pKa = 8.15TAPTLTALQFSALPVVTYY730 pKa = 9.53TYY732 pKa = 11.36DD733 pKa = 3.26VTNSGNVTIPGPITVTDD750 pKa = 4.6DD751 pKa = 3.43KK752 pKa = 11.39LSPQTFTCTGAAIAVGDD769 pKa = 4.35SVQCTATYY777 pKa = 10.35QVTPTDD783 pKa = 3.43VARR786 pKa = 11.84QSVRR790 pKa = 11.84NTAFATGGDD799 pKa = 3.77GTVSNTDD806 pKa = 3.5DD807 pKa = 4.63AVIAPAGRR815 pKa = 11.84LDD817 pKa = 3.5IQLEE821 pKa = 4.29KK822 pKa = 10.79QAVTTDD828 pKa = 4.01FAAVGDD834 pKa = 4.22PVDD837 pKa = 4.0FTLTFTNTGDD847 pKa = 3.36FTLFPEE853 pKa = 4.67TDD855 pKa = 2.97SSALDD860 pKa = 3.51RR861 pKa = 11.84YY862 pKa = 10.18EE863 pKa = 3.86LTDD866 pKa = 4.05PGVSGLDD873 pKa = 3.16SCIIPNPLFPQGGAQTPTQFTCTVTRR899 pKa = 11.84IITQADD905 pKa = 3.34VDD907 pKa = 3.95AGSYY911 pKa = 10.12TNTAFLTMIYY921 pKa = 10.42NQGQVNEE928 pKa = 4.33EE929 pKa = 4.32TQVVPPASDD938 pKa = 3.54TVPLNANVVPSFTLTKK954 pKa = 9.97TATNDD959 pKa = 3.13YY960 pKa = 10.06GAVGDD965 pKa = 4.06TINYY969 pKa = 9.43DD970 pKa = 3.73FEE972 pKa = 4.53VTNTSVLTLTNVVLTDD988 pKa = 3.76PKK990 pKa = 10.24IGSISCVGSATANEE1004 pKa = 4.5TGTIAPGASALCTGSYY1020 pKa = 10.42EE1021 pKa = 4.17VTQADD1026 pKa = 3.85LDD1028 pKa = 4.0AEE1030 pKa = 4.38QVVNTVSALAQTQAAGSVTQTAQEE1054 pKa = 3.93ITPIDD1059 pKa = 3.56PAAATRR1065 pKa = 11.84ALSLTKK1071 pKa = 9.94IVTVRR1076 pKa = 11.84EE1077 pKa = 4.05TGDD1080 pKa = 3.25ATFTAPGQTADD1091 pKa = 3.42YY1092 pKa = 10.46SFVVEE1097 pKa = 4.05NTGNLTLDD1105 pKa = 3.98NIAVVDD1111 pKa = 3.88ADD1113 pKa = 5.17LGYY1116 pKa = 8.65TCQIPTLAPGATDD1129 pKa = 3.28TATCSASRR1137 pKa = 11.84VLTQADD1143 pKa = 4.05FDD1145 pKa = 4.19AGSYY1149 pKa = 10.38QNDD1152 pKa = 3.21AVARR1156 pKa = 11.84AAGADD1161 pKa = 3.9DD1162 pKa = 4.66APASATATAQGRR1174 pKa = 11.84IASFDD1179 pKa = 3.84FVKK1182 pKa = 10.61SAPASFAVAGEE1193 pKa = 4.11VVDD1196 pKa = 5.73FVFAFTNTGTVTLTDD1211 pKa = 3.33ITIVDD1216 pKa = 4.02PLLGVAYY1223 pKa = 9.72ACEE1226 pKa = 4.14ITSIAPGVTDD1236 pKa = 3.68NSCFGSYY1243 pKa = 8.4EE1244 pKa = 4.02VQPADD1249 pKa = 3.25VDD1251 pKa = 3.56AGQIQNDD1258 pKa = 3.37ASFTGVGLDD1267 pKa = 3.62GAPISGDD1274 pKa = 3.1DD1275 pKa = 3.71TVIVTGPAEE1284 pKa = 4.29APALTVTKK1292 pKa = 10.74VEE1294 pKa = 4.53DD1295 pKa = 4.42DD1296 pKa = 3.67GTGTFDD1302 pKa = 4.09NLPATEE1308 pKa = 4.52SYY1310 pKa = 11.29SFVVTNTGNVTLTDD1324 pKa = 4.33LSLVDD1329 pKa = 4.6DD1330 pKa = 4.37LTGFSCALPDD1340 pKa = 4.23LAAGAATSLCADD1352 pKa = 4.69GAPLTDD1358 pKa = 4.1SYY1360 pKa = 11.76TVLQTDD1366 pKa = 3.33IDD1368 pKa = 4.06AGALEE1373 pKa = 4.29NTATATGRR1381 pKa = 11.84TTRR1384 pKa = 11.84GAGVPVEE1391 pKa = 4.64ASDD1394 pKa = 3.75TVTLQGPAQLPVISMVKK1411 pKa = 8.9TATAGAGFSALGDD1424 pKa = 3.88TVSYY1428 pKa = 10.78DD1429 pKa = 3.49YY1430 pKa = 11.34TVTNDD1435 pKa = 3.27GNITLTAPITVTDD1448 pKa = 3.63DD1449 pKa = 3.36RR1450 pKa = 11.84VTVSCPALPAAGLAPGADD1468 pKa = 3.51IVCTATDD1475 pKa = 3.61TVDD1478 pKa = 4.66QADD1481 pKa = 4.04LDD1483 pKa = 3.85AGEE1486 pKa = 4.61IVNTATANVRR1496 pKa = 11.84QPVSPSATYY1505 pKa = 9.4PGGVAEE1511 pKa = 4.32VSSNTEE1517 pKa = 4.15TVTVTADD1524 pKa = 3.37QQPALAITKK1533 pKa = 9.63RR1534 pKa = 11.84VKK1536 pKa = 10.48PGTATSFDD1544 pKa = 3.83APGDD1548 pKa = 3.64LSDD1551 pKa = 5.02PGNPNNLVYY1560 pKa = 10.65EE1561 pKa = 4.85FIVEE1565 pKa = 3.98NTGNVTTTDD1574 pKa = 5.12PITVTDD1580 pKa = 4.45PLISATPLVCTTDD1593 pKa = 3.58PLAPGGTVTCEE1604 pKa = 3.57LPYY1607 pKa = 10.99APTQEE1612 pKa = 4.23DD1613 pKa = 3.26VDD1615 pKa = 3.97RR1616 pKa = 11.84GFFANSADD1624 pKa = 3.17AATRR1628 pKa = 11.84FNGAPVQTPAPGTEE1642 pKa = 4.08TVNAVQRR1649 pKa = 11.84PALTIVKK1656 pKa = 8.64TLEE1659 pKa = 4.05DD1660 pKa = 3.77LAIFSPGEE1668 pKa = 3.38IATYY1672 pKa = 10.34RR1673 pKa = 11.84FYY1675 pKa = 10.89VTNTGNTTITGPVTVTDD1692 pKa = 3.95NKK1694 pKa = 10.1IAGPIDD1700 pKa = 4.37CGPGDD1705 pKa = 4.53LAPGDD1710 pKa = 4.05STACLATYY1718 pKa = 8.42TVTVNDD1724 pKa = 3.58VNAGSVTNSASASNGVTTSDD1744 pKa = 3.95PQSLTIPVGAEE1755 pKa = 3.6PTLSIVKK1762 pKa = 7.61TAPGGVFAQAGDD1774 pKa = 3.71VIAYY1778 pKa = 7.63EE1779 pKa = 4.19FLVTNTSSGPIPPAFVQPVTVIDD1802 pKa = 4.5DD1803 pKa = 4.56KK1804 pKa = 10.78ITTPIVCPVVDD1815 pKa = 4.53GANPLDD1821 pKa = 4.3PGEE1824 pKa = 5.37SITCTGSYY1832 pKa = 8.94TVQQADD1838 pKa = 3.71LDD1840 pKa = 4.0AVRR1843 pKa = 11.84PGFTRR1848 pKa = 11.84GFVINNAFARR1858 pKa = 11.84SEE1860 pKa = 4.04FAGEE1864 pKa = 4.09LVEE1867 pKa = 5.17SPVDD1871 pKa = 3.64QVIITADD1878 pKa = 3.75SVPLLGVVKK1887 pKa = 9.3TAEE1890 pKa = 4.09NQTNPGQDD1898 pKa = 2.86AAAGDD1903 pKa = 4.08VVAFTIVSQNDD1914 pKa = 4.02GPQTLTDD1921 pKa = 3.57VRR1923 pKa = 11.84VTDD1926 pKa = 4.13PFLGALSCDD1935 pKa = 2.94IASPVTLAPGAALTCTGSYY1954 pKa = 9.35TVTQADD1960 pKa = 3.47VDD1962 pKa = 3.87AGGPISNTADD1972 pKa = 3.15AAATTTQGEE1981 pKa = 4.57AVSGTDD1987 pKa = 2.95TTTYY1991 pKa = 10.21PVAAPLPAMQVAKK2004 pKa = 10.5ALEE2007 pKa = 4.6PGSGQNDD2014 pKa = 4.23YY2015 pKa = 9.48NTVGEE2020 pKa = 4.4DD2021 pKa = 3.0LTFRR2025 pKa = 11.84VTVTNTGNITLNDD2038 pKa = 3.49VAVTDD2043 pKa = 4.26DD2044 pKa = 4.99LVPGTCTVPTLAPGASDD2061 pKa = 3.66TSCVFVYY2068 pKa = 9.15TVQQADD2074 pKa = 3.43IDD2076 pKa = 4.05RR2077 pKa = 11.84GTITNIATAQAQPANPGAPEE2097 pKa = 4.85LIEE2100 pKa = 4.45TGQSDD2105 pKa = 3.56ITGPAAEE2112 pKa = 4.87PGVGLSKK2119 pKa = 9.84TGSGEE2124 pKa = 3.92VTASDD2129 pKa = 4.24GSPTFNLPNQSLTYY2143 pKa = 9.93VYY2145 pKa = 10.68EE2146 pKa = 4.11VVNLGNVTLNQLPAITDD2163 pKa = 3.47DD2164 pKa = 4.96RR2165 pKa = 11.84IDD2167 pKa = 4.58SVTCDD2172 pKa = 3.94PLPAAGLLPGEE2183 pKa = 4.32VLICRR2188 pKa = 11.84GSDD2191 pKa = 2.95TVDD2194 pKa = 3.31QNDD2197 pKa = 2.94VDD2199 pKa = 4.46AGFVTNIADD2208 pKa = 3.46VTVANDD2214 pKa = 3.35YY2215 pKa = 11.34GGAPLTAQAQEE2226 pKa = 4.5TITATVAPDD2235 pKa = 3.41LAITKK2240 pKa = 8.44TPSITQNATAGTEE2253 pKa = 3.79VTYY2256 pKa = 10.26SYY2258 pKa = 10.97RR2259 pKa = 11.84VQNPGNVTLTDD2270 pKa = 3.05ITLTDD2275 pKa = 4.01FHH2277 pKa = 6.72TSAAGVVDD2285 pKa = 5.42LPISDD2290 pKa = 4.21GGVIATLAPGEE2301 pKa = 4.35EE2302 pKa = 4.26ATLTATYY2309 pKa = 9.51TVTQDD2314 pKa = 5.55DD2315 pKa = 4.23IDD2317 pKa = 3.83SGNAVRR2323 pKa = 11.84NTVRR2327 pKa = 11.84VTSTLPGGDD2336 pKa = 4.07AGPAPLTRR2344 pKa = 11.84ARR2346 pKa = 11.84VDD2348 pKa = 4.2LDD2350 pKa = 3.38PQRR2353 pKa = 11.84PAMEE2357 pKa = 5.12AIKK2360 pKa = 9.0TIPSSPVPPVPGSQITYY2377 pKa = 10.47QITLTNTGNVTLGDD2391 pKa = 3.9PVLTDD2396 pKa = 3.08TVRR2399 pKa = 11.84RR2400 pKa = 11.84INTDD2404 pKa = 2.94ALAGEE2409 pKa = 4.37PAVALTGGDD2418 pKa = 3.2TGTPGLFEE2426 pKa = 4.23VGEE2429 pKa = 4.16VRR2431 pKa = 11.84TYY2433 pKa = 10.99AVSYY2437 pKa = 9.64TLTQDD2442 pKa = 5.01DD2443 pKa = 3.79IDD2445 pKa = 5.23AGGVANSVTATADD2458 pKa = 3.52DD2459 pKa = 4.26PLGNPVTEE2467 pKa = 4.32VSDD2470 pKa = 3.63NGAGDD2475 pKa = 4.03GDD2477 pKa = 4.56DD2478 pKa = 3.43PTLFVIAPTPALEE2491 pKa = 4.23VIKK2494 pKa = 10.81VISQPASAVGEE2505 pKa = 4.2TVVFEE2510 pKa = 3.96ITATNTGNVTLLDD2523 pKa = 3.83AALTDD2528 pKa = 3.37SFTRR2532 pKa = 11.84LDD2534 pKa = 3.72GTPIPATPP2542 pKa = 3.65
Molecular weight: 259.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q3ESF3|A0A0Q3ESF3_9RHOB Histidinol-phosphate aminotransferase OS=Loktanella sp. 3ANDIMAR09 OX=1225657 GN=AN189_14785 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.35AGRR28 pKa = 11.84NIINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84SKK41 pKa = 11.12LSAA44 pKa = 3.74
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.35AGRR28 pKa = 11.84NIINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84SKK41 pKa = 11.12LSAA44 pKa = 3.74
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1058224 |
32 |
2634 |
300.0 |
32.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.687 ± 0.064 | 0.881 ± 0.012 |
6.887 ± 0.047 | 4.895 ± 0.041 |
3.639 ± 0.028 | 8.679 ± 0.052 |
1.989 ± 0.024 | 5.262 ± 0.031 |
2.798 ± 0.035 | 9.722 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.845 ± 0.024 | 2.636 ± 0.025 |
5.057 ± 0.031 | 3.434 ± 0.024 |
6.688 ± 0.044 | 4.87 ± 0.029 |
6.062 ± 0.043 | 7.421 ± 0.037 |
1.366 ± 0.02 | 2.182 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
