
Human papillomavirus 201
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 27
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
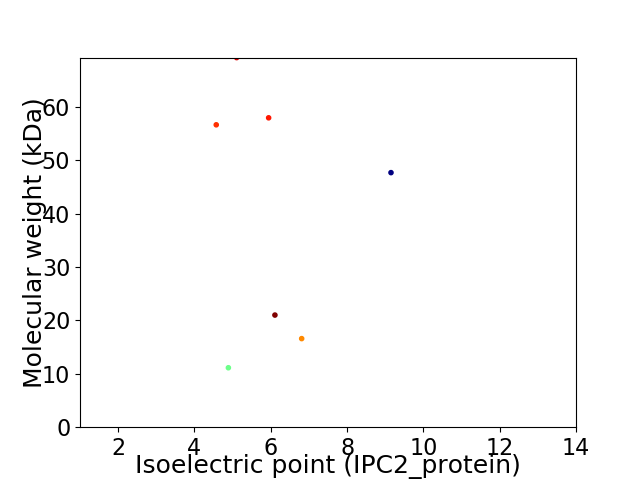
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
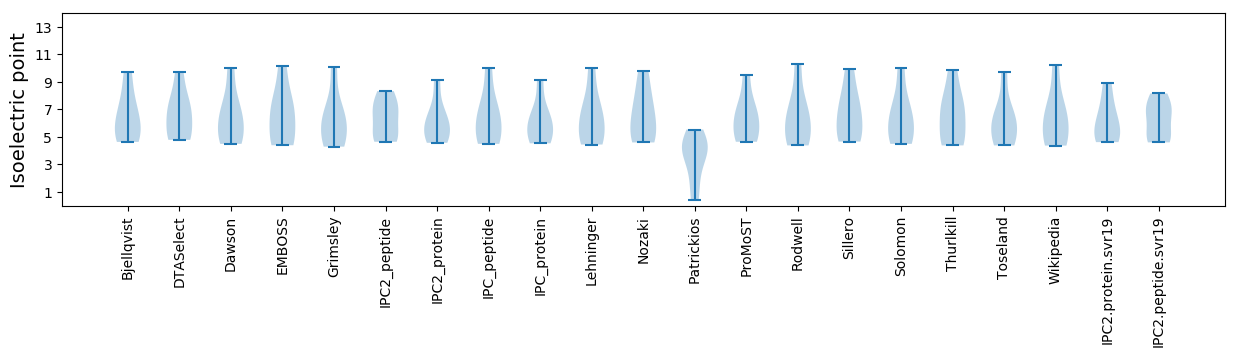
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4LQV7|A0A0H4LQV7_9PAPI Major capsid protein L1 OS=Human papillomavirus 201 OX=1682340 GN=L1 PE=3 SV=1
MM1 pKa = 7.59IGVEE5 pKa = 3.93PTIADD10 pKa = 3.47IEE12 pKa = 4.53LDD14 pKa = 3.74LQEE17 pKa = 5.55LVLPSNLLSNEE28 pKa = 3.92EE29 pKa = 4.07SLEE32 pKa = 4.04EE33 pKa = 3.88QVEE36 pKa = 4.44EE37 pKa = 4.24EE38 pKa = 3.8QQEE41 pKa = 4.77YY42 pKa = 10.44YY43 pKa = 10.71RR44 pKa = 11.84IDD46 pKa = 3.86TRR48 pKa = 11.84CHH50 pKa = 5.43TCGTGVRR57 pKa = 11.84CCVSATSFGIRR68 pKa = 11.84QLEE71 pKa = 3.82LLLIHH76 pKa = 6.72EE77 pKa = 6.0GISFCCPGCSRR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84SFHH93 pKa = 6.54HH94 pKa = 6.91GRR96 pKa = 11.84LQQ98 pKa = 3.32
MM1 pKa = 7.59IGVEE5 pKa = 3.93PTIADD10 pKa = 3.47IEE12 pKa = 4.53LDD14 pKa = 3.74LQEE17 pKa = 5.55LVLPSNLLSNEE28 pKa = 3.92EE29 pKa = 4.07SLEE32 pKa = 4.04EE33 pKa = 3.88QVEE36 pKa = 4.44EE37 pKa = 4.24EE38 pKa = 3.8QQEE41 pKa = 4.77YY42 pKa = 10.44YY43 pKa = 10.71RR44 pKa = 11.84IDD46 pKa = 3.86TRR48 pKa = 11.84CHH50 pKa = 5.43TCGTGVRR57 pKa = 11.84CCVSATSFGIRR68 pKa = 11.84QLEE71 pKa = 3.82LLLIHH76 pKa = 6.72EE77 pKa = 6.0GISFCCPGCSRR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84SFHH93 pKa = 6.54HH94 pKa = 6.91GRR96 pKa = 11.84LQQ98 pKa = 3.32
Molecular weight: 11.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4M3P8|A0A0H4M3P8_9PAPI Regulatory protein E2 OS=Human papillomavirus 201 OX=1682340 GN=E2 PE=3 SV=1
MM1 pKa = 7.71EE2 pKa = 5.0EE3 pKa = 3.62LTRR6 pKa = 11.84RR7 pKa = 11.84FDD9 pKa = 3.57VLQDD13 pKa = 3.1QLMRR17 pKa = 11.84HH18 pKa = 5.61YY19 pKa = 10.29EE20 pKa = 4.14SEE22 pKa = 4.45SKK24 pKa = 10.79SLQDD28 pKa = 4.46QINYY32 pKa = 9.14WEE34 pKa = 4.56TIRR37 pKa = 11.84KK38 pKa = 8.76EE39 pKa = 4.18SALLHH44 pKa = 5.73YY45 pKa = 10.3ARR47 pKa = 11.84QQGISKK53 pKa = 10.5LGLQPVPALVVSEE66 pKa = 4.43YY67 pKa = 10.61NGKK70 pKa = 8.79QAISISLTLQSLLKK84 pKa = 10.77SPFGKK89 pKa = 8.4EE90 pKa = 3.28TWTLPEE96 pKa = 4.16TSAEE100 pKa = 4.46LINTEE105 pKa = 4.71PKK107 pKa = 9.31NTLKK111 pKa = 10.82KK112 pKa = 10.06HH113 pKa = 5.16GFTVTVWFDD122 pKa = 3.39HH123 pKa = 6.17EE124 pKa = 4.58KK125 pKa = 9.93QNAMQYY131 pKa = 8.47TNWNNIYY138 pKa = 10.13YY139 pKa = 9.79QDD141 pKa = 4.43YY142 pKa = 10.51DD143 pKa = 4.2NVWHH147 pKa = 6.38KK148 pKa = 11.39VKK150 pKa = 11.26GEE152 pKa = 3.51VDD154 pKa = 3.52YY155 pKa = 11.8NGLYY159 pKa = 8.66FTEE162 pKa = 4.56PSGDD166 pKa = 2.99RR167 pKa = 11.84AYY169 pKa = 11.04FKK171 pKa = 10.67IFAQDD176 pKa = 2.85AEE178 pKa = 4.78MYY180 pKa = 10.31SKK182 pKa = 9.52TGEE185 pKa = 3.98WTVHH189 pKa = 4.94YY190 pKa = 7.95KK191 pKa = 10.06TYY193 pKa = 10.21TISPPSGFSSASSSYY208 pKa = 11.32SRR210 pKa = 11.84TTEE213 pKa = 3.32ISTSATTRR221 pKa = 11.84HH222 pKa = 5.65TSPEE226 pKa = 3.86KK227 pKa = 10.62ASTSNEE233 pKa = 3.66QQRR236 pKa = 11.84VLRR239 pKa = 11.84SPPRR243 pKa = 11.84LRR245 pKa = 11.84AASVSPPQPSSTTEE259 pKa = 3.67TPEE262 pKa = 3.69RR263 pKa = 11.84EE264 pKa = 3.62RR265 pKa = 11.84VQRR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84RR272 pKa = 11.84EE273 pKa = 3.72QGEE276 pKa = 4.2SRR278 pKa = 11.84PRR280 pKa = 11.84EE281 pKa = 4.03ATPKK285 pKa = 9.96RR286 pKa = 11.84RR287 pKa = 11.84RR288 pKa = 11.84TEE290 pKa = 3.9DD291 pKa = 2.97SGGRR295 pKa = 11.84AGRR298 pKa = 11.84GGGGRR303 pKa = 11.84GAPSPEE309 pKa = 3.64AVGQRR314 pKa = 11.84HH315 pKa = 5.63RR316 pKa = 11.84SVEE319 pKa = 4.28TKK321 pKa = 10.26GLSRR325 pKa = 11.84LRR327 pKa = 11.84ALQEE331 pKa = 3.86EE332 pKa = 4.57ARR334 pKa = 11.84DD335 pKa = 3.65PPVIIIRR342 pKa = 11.84GGANALKK349 pKa = 10.18CFRR352 pKa = 11.84YY353 pKa = 9.99RR354 pKa = 11.84KK355 pKa = 6.75THH357 pKa = 5.99SSSKK361 pKa = 10.66HH362 pKa = 5.16LFLCISSVFKK372 pKa = 10.59WVGDD376 pKa = 3.85CTATSGSRR384 pKa = 11.84MLVSFKK390 pKa = 10.59DD391 pKa = 3.55VKK393 pKa = 9.86QRR395 pKa = 11.84EE396 pKa = 4.02MFIRR400 pKa = 11.84HH401 pKa = 3.96TTFPKK406 pKa = 9.54GSEE409 pKa = 4.2YY410 pKa = 10.74FLGQLNSLL418 pKa = 4.16
MM1 pKa = 7.71EE2 pKa = 5.0EE3 pKa = 3.62LTRR6 pKa = 11.84RR7 pKa = 11.84FDD9 pKa = 3.57VLQDD13 pKa = 3.1QLMRR17 pKa = 11.84HH18 pKa = 5.61YY19 pKa = 10.29EE20 pKa = 4.14SEE22 pKa = 4.45SKK24 pKa = 10.79SLQDD28 pKa = 4.46QINYY32 pKa = 9.14WEE34 pKa = 4.56TIRR37 pKa = 11.84KK38 pKa = 8.76EE39 pKa = 4.18SALLHH44 pKa = 5.73YY45 pKa = 10.3ARR47 pKa = 11.84QQGISKK53 pKa = 10.5LGLQPVPALVVSEE66 pKa = 4.43YY67 pKa = 10.61NGKK70 pKa = 8.79QAISISLTLQSLLKK84 pKa = 10.77SPFGKK89 pKa = 8.4EE90 pKa = 3.28TWTLPEE96 pKa = 4.16TSAEE100 pKa = 4.46LINTEE105 pKa = 4.71PKK107 pKa = 9.31NTLKK111 pKa = 10.82KK112 pKa = 10.06HH113 pKa = 5.16GFTVTVWFDD122 pKa = 3.39HH123 pKa = 6.17EE124 pKa = 4.58KK125 pKa = 9.93QNAMQYY131 pKa = 8.47TNWNNIYY138 pKa = 10.13YY139 pKa = 9.79QDD141 pKa = 4.43YY142 pKa = 10.51DD143 pKa = 4.2NVWHH147 pKa = 6.38KK148 pKa = 11.39VKK150 pKa = 11.26GEE152 pKa = 3.51VDD154 pKa = 3.52YY155 pKa = 11.8NGLYY159 pKa = 8.66FTEE162 pKa = 4.56PSGDD166 pKa = 2.99RR167 pKa = 11.84AYY169 pKa = 11.04FKK171 pKa = 10.67IFAQDD176 pKa = 2.85AEE178 pKa = 4.78MYY180 pKa = 10.31SKK182 pKa = 9.52TGEE185 pKa = 3.98WTVHH189 pKa = 4.94YY190 pKa = 7.95KK191 pKa = 10.06TYY193 pKa = 10.21TISPPSGFSSASSSYY208 pKa = 11.32SRR210 pKa = 11.84TTEE213 pKa = 3.32ISTSATTRR221 pKa = 11.84HH222 pKa = 5.65TSPEE226 pKa = 3.86KK227 pKa = 10.62ASTSNEE233 pKa = 3.66QQRR236 pKa = 11.84VLRR239 pKa = 11.84SPPRR243 pKa = 11.84LRR245 pKa = 11.84AASVSPPQPSSTTEE259 pKa = 3.67TPEE262 pKa = 3.69RR263 pKa = 11.84EE264 pKa = 3.62RR265 pKa = 11.84VQRR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84RR272 pKa = 11.84EE273 pKa = 3.72QGEE276 pKa = 4.2SRR278 pKa = 11.84PRR280 pKa = 11.84EE281 pKa = 4.03ATPKK285 pKa = 9.96RR286 pKa = 11.84RR287 pKa = 11.84RR288 pKa = 11.84TEE290 pKa = 3.9DD291 pKa = 2.97SGGRR295 pKa = 11.84AGRR298 pKa = 11.84GGGGRR303 pKa = 11.84GAPSPEE309 pKa = 3.64AVGQRR314 pKa = 11.84HH315 pKa = 5.63RR316 pKa = 11.84SVEE319 pKa = 4.28TKK321 pKa = 10.26GLSRR325 pKa = 11.84LRR327 pKa = 11.84ALQEE331 pKa = 3.86EE332 pKa = 4.57ARR334 pKa = 11.84DD335 pKa = 3.65PPVIIIRR342 pKa = 11.84GGANALKK349 pKa = 10.18CFRR352 pKa = 11.84YY353 pKa = 9.99RR354 pKa = 11.84KK355 pKa = 6.75THH357 pKa = 5.99SSSKK361 pKa = 10.66HH362 pKa = 5.16LFLCISSVFKK372 pKa = 10.59WVGDD376 pKa = 3.85CTATSGSRR384 pKa = 11.84MLVSFKK390 pKa = 10.59DD391 pKa = 3.55VKK393 pKa = 9.86QRR395 pKa = 11.84EE396 pKa = 4.02MFIRR400 pKa = 11.84HH401 pKa = 3.96TTFPKK406 pKa = 9.54GSEE409 pKa = 4.2YY410 pKa = 10.74FLGQLNSLL418 pKa = 4.16
Molecular weight: 47.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2471 |
98 |
608 |
353.0 |
40.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.261 ± 0.467 | 2.388 ± 0.837 |
5.828 ± 0.732 | 6.92 ± 0.867 |
4.937 ± 0.717 | 5.585 ± 0.479 |
2.185 ± 0.4 | 5.706 ± 0.748 |
5.261 ± 0.838 | 8.822 ± 0.83 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.538 ± 0.389 | 5.059 ± 0.782 |
6.192 ± 0.866 | 4.775 ± 0.188 |
5.949 ± 0.892 | 7.487 ± 0.933 |
6.516 ± 0.787 | 5.221 ± 0.31 |
1.214 ± 0.247 | 3.157 ± 0.422 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
