
Cervus papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Epsilonpapillomavirus; Epsilonpapillomavirus 2
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
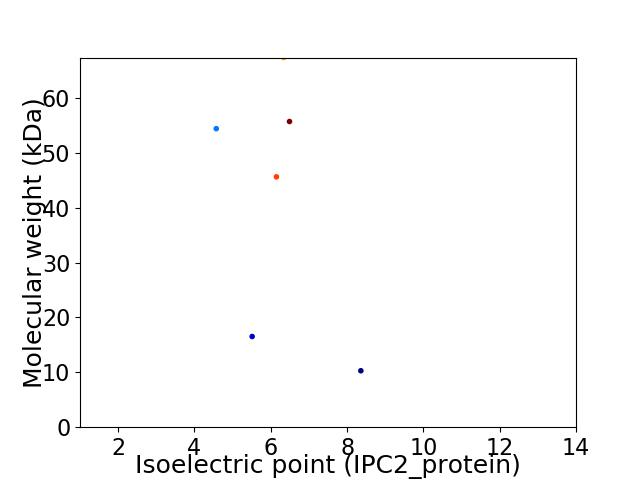
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A182BAF1|A0A182BAF1_9PAPI Minor capsid protein L2 OS=Cervus papillomavirus 2 OX=1434070 GN=L2 PE=3 SV=1
MM1 pKa = 7.49AVRR4 pKa = 11.84VRR6 pKa = 11.84SGRR9 pKa = 11.84VKK11 pKa = 10.4RR12 pKa = 11.84ASPYY16 pKa = 10.89DD17 pKa = 3.77LYY19 pKa = 10.25RR20 pKa = 11.84TCATGDD26 pKa = 3.91CPPDD30 pKa = 3.64VIPKK34 pKa = 9.95VEE36 pKa = 4.17GNTLADD42 pKa = 5.57KK43 pKa = 10.3ILKK46 pKa = 8.85WGSLGVYY53 pKa = 10.42FGGLGIGTGQARR65 pKa = 11.84PGLGTYY71 pKa = 8.92TPLGRR76 pKa = 11.84GGGGVTPRR84 pKa = 11.84PGTRR88 pKa = 11.84PLGTPFGGRR97 pKa = 11.84PIDD100 pKa = 3.96TIGSGIGRR108 pKa = 11.84PGIGRR113 pKa = 11.84PGTIDD118 pKa = 3.49TPVDD122 pKa = 4.0SILPEE127 pKa = 4.21SPAVVTPDD135 pKa = 4.81SMPTDD140 pKa = 4.02PGIGGMDD147 pKa = 3.18VDD149 pKa = 4.13VEE151 pKa = 4.33LVEE154 pKa = 4.43EE155 pKa = 4.17PSIRR159 pKa = 11.84LIEE162 pKa = 3.93PHH164 pKa = 6.97APDD167 pKa = 4.36DD168 pKa = 4.0VAVLDD173 pKa = 4.07VRR175 pKa = 11.84PTEE178 pKa = 4.19HH179 pKa = 7.2DD180 pKa = 3.52LSVRR184 pKa = 11.84TTNSSTVHH192 pKa = 6.34HH193 pKa = 6.57NPSFQGGSTLYY204 pKa = 10.71SEE206 pKa = 4.81VGEE209 pKa = 4.28TSGVEE214 pKa = 3.85NVLIAGSNIGGEE226 pKa = 3.73AHH228 pKa = 7.0EE229 pKa = 4.87NIEE232 pKa = 4.25LQWYY236 pKa = 5.88TEE238 pKa = 4.31PRR240 pKa = 11.84SSTPADD246 pKa = 3.57TPVGRR251 pKa = 11.84VRR253 pKa = 11.84GRR255 pKa = 11.84ANWFSRR261 pKa = 11.84RR262 pKa = 11.84YY263 pKa = 8.06YY264 pKa = 10.44DD265 pKa = 3.7QYY267 pKa = 9.36TAEE270 pKa = 5.55DD271 pKa = 3.71IGLLTDD277 pKa = 3.61PRR279 pKa = 11.84DD280 pKa = 3.43YY281 pKa = 10.62FYY283 pKa = 11.07EE284 pKa = 4.03VEE286 pKa = 4.25NPAFEE291 pKa = 5.16GDD293 pKa = 3.84SFDD296 pKa = 3.62VAGPAVSAEE305 pKa = 3.97PLQVEE310 pKa = 4.67LRR312 pKa = 11.84DD313 pKa = 3.68ITHH316 pKa = 6.11VSAARR321 pKa = 11.84ALSGPSGRR329 pKa = 11.84IGLGRR334 pKa = 11.84IGFRR338 pKa = 11.84STLQTRR344 pKa = 11.84SGVSIGQPLHH354 pKa = 5.65FRR356 pKa = 11.84YY357 pKa = 10.31SFSTIAGEE365 pKa = 4.22TSLEE369 pKa = 3.98LLPHH373 pKa = 6.52SFSQEE378 pKa = 3.8PVDD381 pKa = 3.48VGFYY385 pKa = 7.7EE386 pKa = 4.46TAAGDD391 pKa = 3.75SDD393 pKa = 4.4FVLVDD398 pKa = 4.29LDD400 pKa = 3.98STEE403 pKa = 4.86SIYY406 pKa = 11.14SDD408 pKa = 2.99SHH410 pKa = 8.68LIDD413 pKa = 4.5SDD415 pKa = 3.74TDD417 pKa = 3.74SLHH420 pKa = 6.92GILVLYY426 pKa = 10.0EE427 pKa = 3.98NEE429 pKa = 5.02AIDD432 pKa = 4.01TVPVEE437 pKa = 4.65TVDD440 pKa = 3.99QFPPDD445 pKa = 4.68LIQRR449 pKa = 11.84PVTVPDD455 pKa = 3.18TGLWPSSKK463 pKa = 10.71GEE465 pKa = 3.93PPAVIAEE472 pKa = 4.07DD473 pKa = 5.19DD474 pKa = 3.62IPGIIILTDD483 pKa = 3.08SDD485 pKa = 5.7AGFFWNTFLHH495 pKa = 6.83PSLLSKK501 pKa = 10.85KK502 pKa = 9.86KK503 pKa = 10.4GKK505 pKa = 8.39KK506 pKa = 7.62TFF508 pKa = 3.52
MM1 pKa = 7.49AVRR4 pKa = 11.84VRR6 pKa = 11.84SGRR9 pKa = 11.84VKK11 pKa = 10.4RR12 pKa = 11.84ASPYY16 pKa = 10.89DD17 pKa = 3.77LYY19 pKa = 10.25RR20 pKa = 11.84TCATGDD26 pKa = 3.91CPPDD30 pKa = 3.64VIPKK34 pKa = 9.95VEE36 pKa = 4.17GNTLADD42 pKa = 5.57KK43 pKa = 10.3ILKK46 pKa = 8.85WGSLGVYY53 pKa = 10.42FGGLGIGTGQARR65 pKa = 11.84PGLGTYY71 pKa = 8.92TPLGRR76 pKa = 11.84GGGGVTPRR84 pKa = 11.84PGTRR88 pKa = 11.84PLGTPFGGRR97 pKa = 11.84PIDD100 pKa = 3.96TIGSGIGRR108 pKa = 11.84PGIGRR113 pKa = 11.84PGTIDD118 pKa = 3.49TPVDD122 pKa = 4.0SILPEE127 pKa = 4.21SPAVVTPDD135 pKa = 4.81SMPTDD140 pKa = 4.02PGIGGMDD147 pKa = 3.18VDD149 pKa = 4.13VEE151 pKa = 4.33LVEE154 pKa = 4.43EE155 pKa = 4.17PSIRR159 pKa = 11.84LIEE162 pKa = 3.93PHH164 pKa = 6.97APDD167 pKa = 4.36DD168 pKa = 4.0VAVLDD173 pKa = 4.07VRR175 pKa = 11.84PTEE178 pKa = 4.19HH179 pKa = 7.2DD180 pKa = 3.52LSVRR184 pKa = 11.84TTNSSTVHH192 pKa = 6.34HH193 pKa = 6.57NPSFQGGSTLYY204 pKa = 10.71SEE206 pKa = 4.81VGEE209 pKa = 4.28TSGVEE214 pKa = 3.85NVLIAGSNIGGEE226 pKa = 3.73AHH228 pKa = 7.0EE229 pKa = 4.87NIEE232 pKa = 4.25LQWYY236 pKa = 5.88TEE238 pKa = 4.31PRR240 pKa = 11.84SSTPADD246 pKa = 3.57TPVGRR251 pKa = 11.84VRR253 pKa = 11.84GRR255 pKa = 11.84ANWFSRR261 pKa = 11.84RR262 pKa = 11.84YY263 pKa = 8.06YY264 pKa = 10.44DD265 pKa = 3.7QYY267 pKa = 9.36TAEE270 pKa = 5.55DD271 pKa = 3.71IGLLTDD277 pKa = 3.61PRR279 pKa = 11.84DD280 pKa = 3.43YY281 pKa = 10.62FYY283 pKa = 11.07EE284 pKa = 4.03VEE286 pKa = 4.25NPAFEE291 pKa = 5.16GDD293 pKa = 3.84SFDD296 pKa = 3.62VAGPAVSAEE305 pKa = 3.97PLQVEE310 pKa = 4.67LRR312 pKa = 11.84DD313 pKa = 3.68ITHH316 pKa = 6.11VSAARR321 pKa = 11.84ALSGPSGRR329 pKa = 11.84IGLGRR334 pKa = 11.84IGFRR338 pKa = 11.84STLQTRR344 pKa = 11.84SGVSIGQPLHH354 pKa = 5.65FRR356 pKa = 11.84YY357 pKa = 10.31SFSTIAGEE365 pKa = 4.22TSLEE369 pKa = 3.98LLPHH373 pKa = 6.52SFSQEE378 pKa = 3.8PVDD381 pKa = 3.48VGFYY385 pKa = 7.7EE386 pKa = 4.46TAAGDD391 pKa = 3.75SDD393 pKa = 4.4FVLVDD398 pKa = 4.29LDD400 pKa = 3.98STEE403 pKa = 4.86SIYY406 pKa = 11.14SDD408 pKa = 2.99SHH410 pKa = 8.68LIDD413 pKa = 4.5SDD415 pKa = 3.74TDD417 pKa = 3.74SLHH420 pKa = 6.92GILVLYY426 pKa = 10.0EE427 pKa = 3.98NEE429 pKa = 5.02AIDD432 pKa = 4.01TVPVEE437 pKa = 4.65TVDD440 pKa = 3.99QFPPDD445 pKa = 4.68LIQRR449 pKa = 11.84PVTVPDD455 pKa = 3.18TGLWPSSKK463 pKa = 10.71GEE465 pKa = 3.93PPAVIAEE472 pKa = 4.07DD473 pKa = 5.19DD474 pKa = 3.62IPGIIILTDD483 pKa = 3.08SDD485 pKa = 5.7AGFFWNTFLHH495 pKa = 6.83PSLLSKK501 pKa = 10.85KK502 pKa = 9.86KK503 pKa = 10.4GKK505 pKa = 8.39KK506 pKa = 7.62TFF508 pKa = 3.52
Molecular weight: 54.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A182BAF0|A0A182BAF0_9PAPI Regulatory protein E2 OS=Cervus papillomavirus 2 OX=1434070 GN=E2 PE=3 SV=1
MM1 pKa = 6.99VAGPACLTTLPKK13 pKa = 10.04INKK16 pKa = 8.28PDD18 pKa = 3.53GVEE21 pKa = 4.13LILRR25 pKa = 11.84KK26 pKa = 9.94LPLRR30 pKa = 11.84PPPPKK35 pKa = 9.68PARR38 pKa = 11.84CMLSSSSGIYY48 pKa = 10.44GVFISCICSTTVRR61 pKa = 11.84ICVEE65 pKa = 4.06SGRR68 pKa = 11.84KK69 pKa = 9.27SILSFQDD76 pKa = 3.99LLTNDD81 pKa = 3.99LTFLCPNCARR91 pKa = 11.84KK92 pKa = 10.08NGFF95 pKa = 3.38
MM1 pKa = 6.99VAGPACLTTLPKK13 pKa = 10.04INKK16 pKa = 8.28PDD18 pKa = 3.53GVEE21 pKa = 4.13LILRR25 pKa = 11.84KK26 pKa = 9.94LPLRR30 pKa = 11.84PPPPKK35 pKa = 9.68PARR38 pKa = 11.84CMLSSSSGIYY48 pKa = 10.44GVFISCICSTTVRR61 pKa = 11.84ICVEE65 pKa = 4.06SGRR68 pKa = 11.84KK69 pKa = 9.27SILSFQDD76 pKa = 3.99LLTNDD81 pKa = 3.99LTFLCPNCARR91 pKa = 11.84KK92 pKa = 10.08NGFF95 pKa = 3.38
Molecular weight: 10.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2255 |
95 |
600 |
375.8 |
41.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.253 ± 0.466 | 2.306 ± 0.928 |
6.297 ± 0.441 | 5.765 ± 0.451 |
4.479 ± 0.462 | 7.761 ± 0.992 |
1.818 ± 0.167 | 4.568 ± 0.594 |
4.834 ± 0.853 | 8.47 ± 0.493 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.596 ± 0.257 | 4.479 ± 0.849 |
6.829 ± 1.021 | 3.326 ± 0.601 |
5.942 ± 0.529 | 8.16 ± 0.596 |
6.12 ± 0.732 | 6.563 ± 0.541 |
1.463 ± 0.306 | 2.971 ± 0.251 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
