
Simiduia agarivorans (strain DSM 21679 / JCM 13881 / BCRC 17597 / SA1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Cellvibrionaceae; Simiduia; Simiduia agarivorans
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
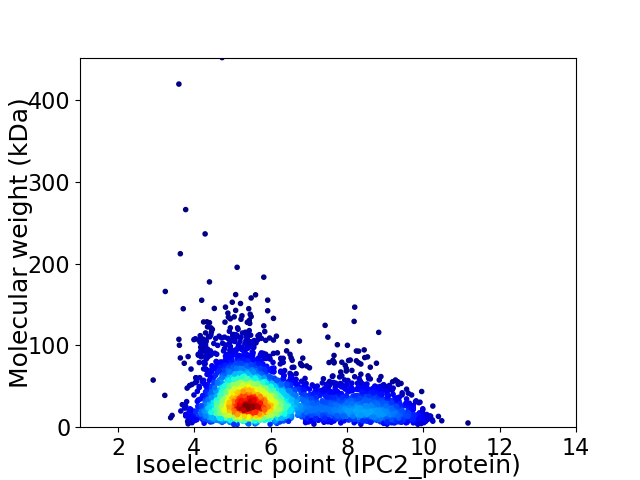
Virtual 2D-PAGE plot for 3812 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4KKL7|K4KKL7_SIMAS rRNA methylase OS=Simiduia agarivorans (strain DSM 21679 / JCM 13881 / BCRC 17597 / SA1) OX=1117647 GN=M5M_12565 PE=4 SV=1
MM1 pKa = 7.35SGWNTSVAGKK11 pKa = 10.16LLLSTSISLLVACGGGGGGAPAPTPTPAPTPAPSPTPTPTPTPTPEE57 pKa = 4.05PTPTPTPTPTPTPEE71 pKa = 3.98PTPTPEE77 pKa = 3.98PTPTPEE83 pKa = 4.08PTPTPTPTPTPTPEE97 pKa = 4.05PTPTPTPTPTPTPTGFAVSGSVSGLNNGYY126 pKa = 7.93YY127 pKa = 10.22TLTLNGTEE135 pKa = 4.3SVTVEE140 pKa = 3.98ANASSFSFQTLLSTDD155 pKa = 2.78ASYY158 pKa = 11.06QVVASGEE165 pKa = 4.16NSTQFCTLAGGEE177 pKa = 4.39GVIGAANVSNLSLSCFGANGYY198 pKa = 6.79TLQGYY203 pKa = 7.72GLQTQSPSIVVMGLRR218 pKa = 11.84VTDD221 pKa = 4.39LASGSPAPEE230 pKa = 3.72NSFSDD235 pKa = 3.14SHH237 pKa = 6.84FVVKK241 pKa = 10.68EE242 pKa = 3.87NGSSVGSEE250 pKa = 3.98SFISAEE256 pKa = 4.01PVANAALDD264 pKa = 3.75LRR266 pKa = 11.84TVIVLDD272 pKa = 3.99ISQSLTPTDD281 pKa = 3.58VEE283 pKa = 4.74DD284 pKa = 3.57IKK286 pKa = 11.44VAAKK290 pKa = 9.99QAIFTTNGDD299 pKa = 3.29GSKK302 pKa = 9.67TSNLVPGQRR311 pKa = 11.84VAIYY315 pKa = 9.64TFDD318 pKa = 3.6STVKK322 pKa = 10.33LVSDD326 pKa = 4.52FSDD329 pKa = 5.47DD330 pKa = 3.17ITALDD335 pKa = 3.72QQIDD339 pKa = 4.3SIDD342 pKa = 3.46SSLVNRR348 pKa = 11.84GNSTNLYY355 pKa = 10.21GAIQTAVGRR364 pKa = 11.84WTNSFTLTSAQFGYY378 pKa = 9.47TIVITDD384 pKa = 3.69GDD386 pKa = 4.23HH387 pKa = 6.59NADD390 pKa = 3.3SRR392 pKa = 11.84TAADD396 pKa = 3.64ISASTTGKK404 pKa = 10.23DD405 pKa = 2.72IYY407 pKa = 10.84AIAVGGDD414 pKa = 3.17VTLANLEE421 pKa = 4.27QVTGSADD428 pKa = 3.45RR429 pKa = 11.84VFTVADD435 pKa = 3.55IGALNAQLDD444 pKa = 4.79EE445 pKa = 4.34IQQAAIDD452 pKa = 3.97QTKK455 pKa = 9.13GLYY458 pKa = 9.45RR459 pKa = 11.84VFYY462 pKa = 8.26ATPKK466 pKa = 9.97RR467 pKa = 11.84SGTHH471 pKa = 5.14EE472 pKa = 4.63VILSLSPEE480 pKa = 4.31YY481 pKa = 10.72TCDD484 pKa = 3.5AQISGCITGLSGTFNSNGFTDD505 pKa = 5.23VIPEE509 pKa = 4.25IYY511 pKa = 10.23VSHH514 pKa = 7.71DD515 pKa = 2.93KK516 pKa = 11.29GKK518 pKa = 10.38EE519 pKa = 3.68VSTNNWAVTPDD530 pKa = 3.23TDD532 pKa = 3.45ITLTSKK538 pKa = 10.64LRR540 pKa = 11.84WANITPAFGYY550 pKa = 9.67SWSNTSGAEE559 pKa = 3.85ATLTAIEE566 pKa = 4.38GTNQQVLNVGLDD578 pKa = 4.01FVQSDD583 pKa = 4.12LLIADD588 pKa = 4.0SASGLSRR595 pKa = 11.84TITYY599 pKa = 10.41HH600 pKa = 7.34LDD602 pKa = 3.23TDD604 pKa = 3.93GDD606 pKa = 4.55GIIDD610 pKa = 3.83TTDD613 pKa = 3.05TDD615 pKa = 4.06DD616 pKa = 6.11DD617 pKa = 4.52NDD619 pKa = 3.95GVLDD623 pKa = 3.67VDD625 pKa = 4.86DD626 pKa = 5.9EE627 pKa = 4.34MRR629 pKa = 11.84QDD631 pKa = 3.43PSEE634 pKa = 4.08TLDD637 pKa = 3.47TDD639 pKa = 3.86GDD641 pKa = 4.61GIGNNADD648 pKa = 3.63TDD650 pKa = 4.11DD651 pKa = 5.5DD652 pKa = 5.15GDD654 pKa = 4.09GVADD658 pKa = 3.83VDD660 pKa = 5.33DD661 pKa = 5.88AFPLDD666 pKa = 3.82NQEE669 pKa = 5.11SLDD672 pKa = 3.78TDD674 pKa = 3.68GDD676 pKa = 4.57GIGNNADD683 pKa = 4.0PDD685 pKa = 4.17DD686 pKa = 5.9DD687 pKa = 4.52NDD689 pKa = 4.0GVADD693 pKa = 4.24EE694 pKa = 5.23SDD696 pKa = 4.22AFPLDD701 pKa = 3.43ASEE704 pKa = 4.35TTDD707 pKa = 3.24TDD709 pKa = 3.54GDD711 pKa = 4.53GIGDD715 pKa = 3.68NADD718 pKa = 3.48PDD720 pKa = 4.11KK721 pKa = 11.09DD722 pKa = 3.9VPSAMGEE729 pKa = 4.35SYY731 pKa = 10.24TLDD734 pKa = 3.37TFAVTPAGALLNVLSNDD751 pKa = 3.24TFGLDD756 pKa = 3.62GAGSLSIAVQPSHH769 pKa = 6.79GSVSLDD775 pKa = 3.75DD776 pKa = 4.44NGTPEE781 pKa = 5.27DD782 pKa = 4.3ASDD785 pKa = 3.58DD786 pKa = 3.65QLRR789 pKa = 11.84FIPADD794 pKa = 3.49GYY796 pKa = 10.39MGSDD800 pKa = 2.73QFTYY804 pKa = 10.53EE805 pKa = 3.92IADD808 pKa = 3.74GSGSTTQALVDD819 pKa = 3.82LTIGGIRR826 pKa = 11.84SLTVSASEE834 pKa = 3.57KK835 pKa = 9.75HH836 pKa = 4.92GQIRR840 pKa = 11.84VSWDD844 pKa = 3.24AQPGANVSHH853 pKa = 5.94YY854 pKa = 9.16TLLVNPDD861 pKa = 3.32GASGYY866 pKa = 8.67TPVAGAEE873 pKa = 4.04MLLVDD878 pKa = 4.51TIYY881 pKa = 11.32YY882 pKa = 10.03DD883 pKa = 3.58IEE885 pKa = 4.59VSVLNYY891 pKa = 10.93DD892 pKa = 4.99FINASYY898 pKa = 9.82MLEE901 pKa = 4.07LRR903 pKa = 11.84DD904 pKa = 3.48SSEE907 pKa = 3.98NVLEE911 pKa = 5.1DD912 pKa = 3.82GLVLVNGLSSSDD924 pKa = 3.01LVTYY928 pKa = 10.63IKK930 pKa = 10.75ASNTDD935 pKa = 2.79AGDD938 pKa = 3.51RR939 pKa = 11.84FGSSVSVSNDD949 pKa = 2.6GTVIAVGAKK958 pKa = 10.29FEE960 pKa = 4.68DD961 pKa = 3.95SAGNGVYY968 pKa = 10.97GNISAGNSSFTDD980 pKa = 3.05SGAVYY985 pKa = 10.35VYY987 pKa = 11.15SKK989 pKa = 11.48LNGVWQQQAYY999 pKa = 9.17IKK1001 pKa = 10.86APLASGASDD1010 pKa = 3.35FFGYY1014 pKa = 9.97SVSLSGDD1021 pKa = 3.11GSTLAVGAYY1030 pKa = 10.3GEE1032 pKa = 4.58DD1033 pKa = 3.88SEE1035 pKa = 5.14LTGSNAPYY1043 pKa = 10.72NNTGTDD1049 pKa = 2.93SGAVYY1054 pKa = 10.51IFTQNAGVWASQAYY1068 pKa = 10.07LKK1070 pKa = 10.36AQNIGHH1076 pKa = 6.48YY1077 pKa = 9.68DD1078 pKa = 3.37YY1079 pKa = 11.39FGFSVSLSTDD1089 pKa = 3.24GNVLAVGAYY1098 pKa = 10.1NEE1100 pKa = 4.86GGSATGVNGVVDD1112 pKa = 3.93NLAAGSGAAYY1122 pKa = 10.22VFNRR1126 pKa = 11.84DD1127 pKa = 3.49GINWSQSAYY1136 pKa = 10.37VKK1138 pKa = 10.81ASNTDD1143 pKa = 3.08ASDD1146 pKa = 3.21MFGFSVGLSGDD1157 pKa = 3.47GNTLVVGAPGEE1168 pKa = 4.68DD1169 pKa = 3.76GASTGIYY1176 pKa = 10.41SVDD1179 pKa = 3.13QSSNGKK1185 pKa = 8.97SSSGAVYY1192 pKa = 9.82VFNYY1196 pKa = 10.05EE1197 pKa = 3.8GTEE1200 pKa = 4.17WVQDD1204 pKa = 3.77SYY1206 pKa = 12.22VKK1208 pKa = 10.95ANNTDD1213 pKa = 3.17SDD1215 pKa = 3.96DD1216 pKa = 3.63QFGYY1220 pKa = 10.04SVSVSADD1227 pKa = 2.88GRR1229 pKa = 11.84DD1230 pKa = 3.64FAVGAIGEE1238 pKa = 4.4DD1239 pKa = 3.12WSGQNPINNNVSNSGAVFMFKK1260 pKa = 9.34KK1261 pKa = 6.8TTYY1264 pKa = 8.93WYY1266 pKa = 10.44QWDD1269 pKa = 3.9HH1270 pKa = 6.46LKK1272 pKa = 11.05ASTPAAKK1279 pKa = 10.43DD1280 pKa = 3.17NFGFSVSISPDD1291 pKa = 2.92GKK1293 pKa = 9.58SVVVGSVFEE1302 pKa = 4.36ASNSVGINPIDD1313 pKa = 3.61EE1314 pKa = 4.72NNFNAQRR1321 pKa = 11.84GAAYY1325 pKa = 10.0LFRR1328 pKa = 11.84SSEE1331 pKa = 4.23PNDD1334 pKa = 3.12HH1335 pKa = 7.51WIQTHH1340 pKa = 5.75YY1341 pKa = 10.41IKK1343 pKa = 10.86SPHH1346 pKa = 5.93TGSNITLGRR1355 pKa = 11.84SVAVSDD1361 pKa = 3.72NANTIVIGADD1371 pKa = 3.66GEE1373 pKa = 4.28NSAATGINSTPTGSKK1388 pKa = 7.6TASGAVYY1395 pKa = 10.11VYY1397 pKa = 10.76RR1398 pKa = 5.83
MM1 pKa = 7.35SGWNTSVAGKK11 pKa = 10.16LLLSTSISLLVACGGGGGGAPAPTPTPAPTPAPSPTPTPTPTPTPEE57 pKa = 4.05PTPTPTPTPTPTPEE71 pKa = 3.98PTPTPEE77 pKa = 3.98PTPTPEE83 pKa = 4.08PTPTPTPTPTPTPEE97 pKa = 4.05PTPTPTPTPTPTPTGFAVSGSVSGLNNGYY126 pKa = 7.93YY127 pKa = 10.22TLTLNGTEE135 pKa = 4.3SVTVEE140 pKa = 3.98ANASSFSFQTLLSTDD155 pKa = 2.78ASYY158 pKa = 11.06QVVASGEE165 pKa = 4.16NSTQFCTLAGGEE177 pKa = 4.39GVIGAANVSNLSLSCFGANGYY198 pKa = 6.79TLQGYY203 pKa = 7.72GLQTQSPSIVVMGLRR218 pKa = 11.84VTDD221 pKa = 4.39LASGSPAPEE230 pKa = 3.72NSFSDD235 pKa = 3.14SHH237 pKa = 6.84FVVKK241 pKa = 10.68EE242 pKa = 3.87NGSSVGSEE250 pKa = 3.98SFISAEE256 pKa = 4.01PVANAALDD264 pKa = 3.75LRR266 pKa = 11.84TVIVLDD272 pKa = 3.99ISQSLTPTDD281 pKa = 3.58VEE283 pKa = 4.74DD284 pKa = 3.57IKK286 pKa = 11.44VAAKK290 pKa = 9.99QAIFTTNGDD299 pKa = 3.29GSKK302 pKa = 9.67TSNLVPGQRR311 pKa = 11.84VAIYY315 pKa = 9.64TFDD318 pKa = 3.6STVKK322 pKa = 10.33LVSDD326 pKa = 4.52FSDD329 pKa = 5.47DD330 pKa = 3.17ITALDD335 pKa = 3.72QQIDD339 pKa = 4.3SIDD342 pKa = 3.46SSLVNRR348 pKa = 11.84GNSTNLYY355 pKa = 10.21GAIQTAVGRR364 pKa = 11.84WTNSFTLTSAQFGYY378 pKa = 9.47TIVITDD384 pKa = 3.69GDD386 pKa = 4.23HH387 pKa = 6.59NADD390 pKa = 3.3SRR392 pKa = 11.84TAADD396 pKa = 3.64ISASTTGKK404 pKa = 10.23DD405 pKa = 2.72IYY407 pKa = 10.84AIAVGGDD414 pKa = 3.17VTLANLEE421 pKa = 4.27QVTGSADD428 pKa = 3.45RR429 pKa = 11.84VFTVADD435 pKa = 3.55IGALNAQLDD444 pKa = 4.79EE445 pKa = 4.34IQQAAIDD452 pKa = 3.97QTKK455 pKa = 9.13GLYY458 pKa = 9.45RR459 pKa = 11.84VFYY462 pKa = 8.26ATPKK466 pKa = 9.97RR467 pKa = 11.84SGTHH471 pKa = 5.14EE472 pKa = 4.63VILSLSPEE480 pKa = 4.31YY481 pKa = 10.72TCDD484 pKa = 3.5AQISGCITGLSGTFNSNGFTDD505 pKa = 5.23VIPEE509 pKa = 4.25IYY511 pKa = 10.23VSHH514 pKa = 7.71DD515 pKa = 2.93KK516 pKa = 11.29GKK518 pKa = 10.38EE519 pKa = 3.68VSTNNWAVTPDD530 pKa = 3.23TDD532 pKa = 3.45ITLTSKK538 pKa = 10.64LRR540 pKa = 11.84WANITPAFGYY550 pKa = 9.67SWSNTSGAEE559 pKa = 3.85ATLTAIEE566 pKa = 4.38GTNQQVLNVGLDD578 pKa = 4.01FVQSDD583 pKa = 4.12LLIADD588 pKa = 4.0SASGLSRR595 pKa = 11.84TITYY599 pKa = 10.41HH600 pKa = 7.34LDD602 pKa = 3.23TDD604 pKa = 3.93GDD606 pKa = 4.55GIIDD610 pKa = 3.83TTDD613 pKa = 3.05TDD615 pKa = 4.06DD616 pKa = 6.11DD617 pKa = 4.52NDD619 pKa = 3.95GVLDD623 pKa = 3.67VDD625 pKa = 4.86DD626 pKa = 5.9EE627 pKa = 4.34MRR629 pKa = 11.84QDD631 pKa = 3.43PSEE634 pKa = 4.08TLDD637 pKa = 3.47TDD639 pKa = 3.86GDD641 pKa = 4.61GIGNNADD648 pKa = 3.63TDD650 pKa = 4.11DD651 pKa = 5.5DD652 pKa = 5.15GDD654 pKa = 4.09GVADD658 pKa = 3.83VDD660 pKa = 5.33DD661 pKa = 5.88AFPLDD666 pKa = 3.82NQEE669 pKa = 5.11SLDD672 pKa = 3.78TDD674 pKa = 3.68GDD676 pKa = 4.57GIGNNADD683 pKa = 4.0PDD685 pKa = 4.17DD686 pKa = 5.9DD687 pKa = 4.52NDD689 pKa = 4.0GVADD693 pKa = 4.24EE694 pKa = 5.23SDD696 pKa = 4.22AFPLDD701 pKa = 3.43ASEE704 pKa = 4.35TTDD707 pKa = 3.24TDD709 pKa = 3.54GDD711 pKa = 4.53GIGDD715 pKa = 3.68NADD718 pKa = 3.48PDD720 pKa = 4.11KK721 pKa = 11.09DD722 pKa = 3.9VPSAMGEE729 pKa = 4.35SYY731 pKa = 10.24TLDD734 pKa = 3.37TFAVTPAGALLNVLSNDD751 pKa = 3.24TFGLDD756 pKa = 3.62GAGSLSIAVQPSHH769 pKa = 6.79GSVSLDD775 pKa = 3.75DD776 pKa = 4.44NGTPEE781 pKa = 5.27DD782 pKa = 4.3ASDD785 pKa = 3.58DD786 pKa = 3.65QLRR789 pKa = 11.84FIPADD794 pKa = 3.49GYY796 pKa = 10.39MGSDD800 pKa = 2.73QFTYY804 pKa = 10.53EE805 pKa = 3.92IADD808 pKa = 3.74GSGSTTQALVDD819 pKa = 3.82LTIGGIRR826 pKa = 11.84SLTVSASEE834 pKa = 3.57KK835 pKa = 9.75HH836 pKa = 4.92GQIRR840 pKa = 11.84VSWDD844 pKa = 3.24AQPGANVSHH853 pKa = 5.94YY854 pKa = 9.16TLLVNPDD861 pKa = 3.32GASGYY866 pKa = 8.67TPVAGAEE873 pKa = 4.04MLLVDD878 pKa = 4.51TIYY881 pKa = 11.32YY882 pKa = 10.03DD883 pKa = 3.58IEE885 pKa = 4.59VSVLNYY891 pKa = 10.93DD892 pKa = 4.99FINASYY898 pKa = 9.82MLEE901 pKa = 4.07LRR903 pKa = 11.84DD904 pKa = 3.48SSEE907 pKa = 3.98NVLEE911 pKa = 5.1DD912 pKa = 3.82GLVLVNGLSSSDD924 pKa = 3.01LVTYY928 pKa = 10.63IKK930 pKa = 10.75ASNTDD935 pKa = 2.79AGDD938 pKa = 3.51RR939 pKa = 11.84FGSSVSVSNDD949 pKa = 2.6GTVIAVGAKK958 pKa = 10.29FEE960 pKa = 4.68DD961 pKa = 3.95SAGNGVYY968 pKa = 10.97GNISAGNSSFTDD980 pKa = 3.05SGAVYY985 pKa = 10.35VYY987 pKa = 11.15SKK989 pKa = 11.48LNGVWQQQAYY999 pKa = 9.17IKK1001 pKa = 10.86APLASGASDD1010 pKa = 3.35FFGYY1014 pKa = 9.97SVSLSGDD1021 pKa = 3.11GSTLAVGAYY1030 pKa = 10.3GEE1032 pKa = 4.58DD1033 pKa = 3.88SEE1035 pKa = 5.14LTGSNAPYY1043 pKa = 10.72NNTGTDD1049 pKa = 2.93SGAVYY1054 pKa = 10.51IFTQNAGVWASQAYY1068 pKa = 10.07LKK1070 pKa = 10.36AQNIGHH1076 pKa = 6.48YY1077 pKa = 9.68DD1078 pKa = 3.37YY1079 pKa = 11.39FGFSVSLSTDD1089 pKa = 3.24GNVLAVGAYY1098 pKa = 10.1NEE1100 pKa = 4.86GGSATGVNGVVDD1112 pKa = 3.93NLAAGSGAAYY1122 pKa = 10.22VFNRR1126 pKa = 11.84DD1127 pKa = 3.49GINWSQSAYY1136 pKa = 10.37VKK1138 pKa = 10.81ASNTDD1143 pKa = 3.08ASDD1146 pKa = 3.21MFGFSVGLSGDD1157 pKa = 3.47GNTLVVGAPGEE1168 pKa = 4.68DD1169 pKa = 3.76GASTGIYY1176 pKa = 10.41SVDD1179 pKa = 3.13QSSNGKK1185 pKa = 8.97SSSGAVYY1192 pKa = 9.82VFNYY1196 pKa = 10.05EE1197 pKa = 3.8GTEE1200 pKa = 4.17WVQDD1204 pKa = 3.77SYY1206 pKa = 12.22VKK1208 pKa = 10.95ANNTDD1213 pKa = 3.17SDD1215 pKa = 3.96DD1216 pKa = 3.63QFGYY1220 pKa = 10.04SVSVSADD1227 pKa = 2.88GRR1229 pKa = 11.84DD1230 pKa = 3.64FAVGAIGEE1238 pKa = 4.4DD1239 pKa = 3.12WSGQNPINNNVSNSGAVFMFKK1260 pKa = 9.34KK1261 pKa = 6.8TTYY1264 pKa = 8.93WYY1266 pKa = 10.44QWDD1269 pKa = 3.9HH1270 pKa = 6.46LKK1272 pKa = 11.05ASTPAAKK1279 pKa = 10.43DD1280 pKa = 3.17NFGFSVSISPDD1291 pKa = 2.92GKK1293 pKa = 9.58SVVVGSVFEE1302 pKa = 4.36ASNSVGINPIDD1313 pKa = 3.61EE1314 pKa = 4.72NNFNAQRR1321 pKa = 11.84GAAYY1325 pKa = 10.0LFRR1328 pKa = 11.84SSEE1331 pKa = 4.23PNDD1334 pKa = 3.12HH1335 pKa = 7.51WIQTHH1340 pKa = 5.75YY1341 pKa = 10.41IKK1343 pKa = 10.86SPHH1346 pKa = 5.93TGSNITLGRR1355 pKa = 11.84SVAVSDD1361 pKa = 3.72NANTIVIGADD1371 pKa = 3.66GEE1373 pKa = 4.28NSAATGINSTPTGSKK1388 pKa = 7.6TASGAVYY1395 pKa = 10.11VYY1397 pKa = 10.76RR1398 pKa = 5.83
Molecular weight: 144.86 kDa
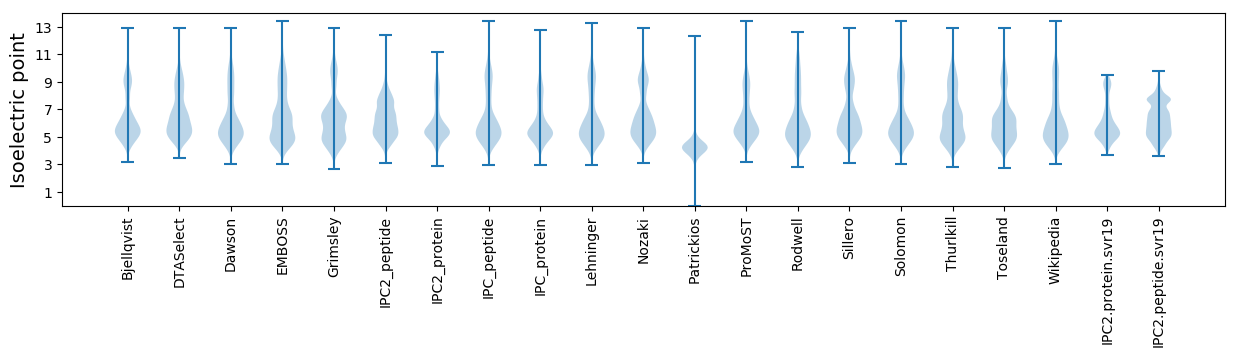
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4L0S2|K4L0S2_SIMAS Probable membrane transporter protein OS=Simiduia agarivorans (strain DSM 21679 / JCM 13881 / BCRC 17597 / SA1) OX=1117647 GN=M5M_12985 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 9.74RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.25GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.12GRR39 pKa = 11.84KK40 pKa = 7.34QLSVAA45 pKa = 3.87
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 9.74RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.25GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.12GRR39 pKa = 11.84KK40 pKa = 7.34QLSVAA45 pKa = 3.87
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1294979 |
29 |
4336 |
339.7 |
37.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.91 ± 0.053 | 1.035 ± 0.013 |
5.784 ± 0.027 | 5.656 ± 0.044 |
3.815 ± 0.026 | 7.589 ± 0.052 |
2.27 ± 0.016 | 5.004 ± 0.031 |
3.848 ± 0.036 | 10.848 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.37 ± 0.02 | 3.636 ± 0.036 |
4.55 ± 0.03 | 4.627 ± 0.03 |
5.602 ± 0.039 | 5.914 ± 0.038 |
5.2 ± 0.038 | 7.028 ± 0.035 |
1.488 ± 0.019 | 2.828 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
