
Wenzhou tombus-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
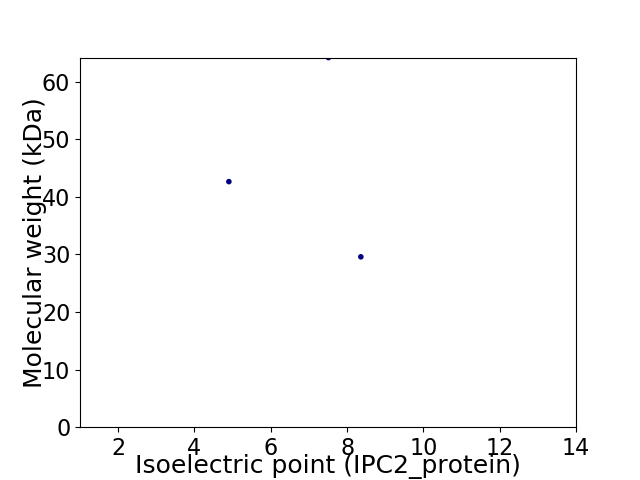
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
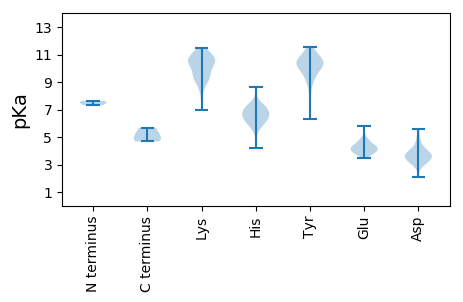
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH35|A0A1L3KH35_9VIRU Uncharacterized protein OS=Wenzhou tombus-like virus 9 OX=1923679 PE=4 SV=1
MM1 pKa = 7.31GMSNTSSSPNMQGQRR16 pKa = 11.84DD17 pKa = 3.46GSTRR21 pKa = 11.84VRR23 pKa = 11.84HH24 pKa = 6.03RR25 pKa = 11.84EE26 pKa = 3.76FITNVGAEE34 pKa = 4.19FGFDD38 pKa = 2.99TKK40 pKa = 11.12LYY42 pKa = 6.33TTGINPGDD50 pKa = 3.75GKK52 pKa = 9.09TFPWLSQLAKK62 pKa = 10.21GYY64 pKa = 9.91EE65 pKa = 4.11RR66 pKa = 11.84YY67 pKa = 9.39HH68 pKa = 6.15VNSMRR73 pKa = 11.84FEE75 pKa = 4.18YY76 pKa = 10.73EE77 pKa = 3.78PFSATTTSGTVSLLVDD93 pKa = 4.36YY94 pKa = 10.9DD95 pKa = 3.73PTDD98 pKa = 4.06PGPRR102 pKa = 11.84SKK104 pKa = 11.15SEE106 pKa = 3.83LLNSYY111 pKa = 9.98RR112 pKa = 11.84AVRR115 pKa = 11.84SSIWMSSHH123 pKa = 6.89TILSKK128 pKa = 11.47AEE130 pKa = 4.18LDD132 pKa = 3.83RR133 pKa = 11.84DD134 pKa = 3.36DD135 pKa = 4.7HH136 pKa = 7.34LFVRR140 pKa = 11.84TTSRR144 pKa = 11.84DD145 pKa = 3.44LMSEE149 pKa = 3.8NLKK152 pKa = 10.89LYY154 pKa = 10.64DD155 pKa = 4.16VGTLFCAFTDD165 pKa = 3.46IADD168 pKa = 3.34VSTLHH173 pKa = 6.49GEE175 pKa = 3.95LWVSYY180 pKa = 9.97DD181 pKa = 3.18ITLMIPSFHH190 pKa = 6.92LSSINNAEE198 pKa = 3.99VNALIVTDD206 pKa = 3.53SRR208 pKa = 11.84YY209 pKa = 10.21LGRR212 pKa = 11.84IDD214 pKa = 4.58PDD216 pKa = 3.28STANEE221 pKa = 4.3KK222 pKa = 10.74GSSLSYY228 pKa = 9.37STHH231 pKa = 6.52QIDD234 pKa = 3.66DD235 pKa = 3.3NAYY238 pKa = 9.19IEE240 pKa = 4.38FNEE243 pKa = 4.41PFSGMVQVSTRR254 pKa = 11.84TVSGDD259 pKa = 3.27LFQIEE264 pKa = 4.68PTLSPAAPISRR275 pKa = 11.84LAKK278 pKa = 9.63IGNAVSYY285 pKa = 9.11FVSNIDD291 pKa = 3.21TWLHH295 pKa = 4.18TVEE298 pKa = 4.56VVADD302 pKa = 3.85AGEE305 pKa = 4.0RR306 pKa = 11.84LAWRR310 pKa = 11.84IVDD313 pKa = 3.96QTAAWAGSLDD323 pKa = 4.26LLFTEE328 pKa = 4.78YY329 pKa = 11.11APALMGTLLALRR341 pKa = 11.84TDD343 pKa = 3.81EE344 pKa = 5.24AEE346 pKa = 3.91LTARR350 pKa = 11.84LQSDD354 pKa = 3.59HH355 pKa = 7.17LIGRR359 pKa = 11.84LPAPSSWTDD368 pKa = 2.11WRR370 pKa = 11.84TQHH373 pKa = 7.05GIPNSTTTEE382 pKa = 3.75THH384 pKa = 5.68
MM1 pKa = 7.31GMSNTSSSPNMQGQRR16 pKa = 11.84DD17 pKa = 3.46GSTRR21 pKa = 11.84VRR23 pKa = 11.84HH24 pKa = 6.03RR25 pKa = 11.84EE26 pKa = 3.76FITNVGAEE34 pKa = 4.19FGFDD38 pKa = 2.99TKK40 pKa = 11.12LYY42 pKa = 6.33TTGINPGDD50 pKa = 3.75GKK52 pKa = 9.09TFPWLSQLAKK62 pKa = 10.21GYY64 pKa = 9.91EE65 pKa = 4.11RR66 pKa = 11.84YY67 pKa = 9.39HH68 pKa = 6.15VNSMRR73 pKa = 11.84FEE75 pKa = 4.18YY76 pKa = 10.73EE77 pKa = 3.78PFSATTTSGTVSLLVDD93 pKa = 4.36YY94 pKa = 10.9DD95 pKa = 3.73PTDD98 pKa = 4.06PGPRR102 pKa = 11.84SKK104 pKa = 11.15SEE106 pKa = 3.83LLNSYY111 pKa = 9.98RR112 pKa = 11.84AVRR115 pKa = 11.84SSIWMSSHH123 pKa = 6.89TILSKK128 pKa = 11.47AEE130 pKa = 4.18LDD132 pKa = 3.83RR133 pKa = 11.84DD134 pKa = 3.36DD135 pKa = 4.7HH136 pKa = 7.34LFVRR140 pKa = 11.84TTSRR144 pKa = 11.84DD145 pKa = 3.44LMSEE149 pKa = 3.8NLKK152 pKa = 10.89LYY154 pKa = 10.64DD155 pKa = 4.16VGTLFCAFTDD165 pKa = 3.46IADD168 pKa = 3.34VSTLHH173 pKa = 6.49GEE175 pKa = 3.95LWVSYY180 pKa = 9.97DD181 pKa = 3.18ITLMIPSFHH190 pKa = 6.92LSSINNAEE198 pKa = 3.99VNALIVTDD206 pKa = 3.53SRR208 pKa = 11.84YY209 pKa = 10.21LGRR212 pKa = 11.84IDD214 pKa = 4.58PDD216 pKa = 3.28STANEE221 pKa = 4.3KK222 pKa = 10.74GSSLSYY228 pKa = 9.37STHH231 pKa = 6.52QIDD234 pKa = 3.66DD235 pKa = 3.3NAYY238 pKa = 9.19IEE240 pKa = 4.38FNEE243 pKa = 4.41PFSGMVQVSTRR254 pKa = 11.84TVSGDD259 pKa = 3.27LFQIEE264 pKa = 4.68PTLSPAAPISRR275 pKa = 11.84LAKK278 pKa = 9.63IGNAVSYY285 pKa = 9.11FVSNIDD291 pKa = 3.21TWLHH295 pKa = 4.18TVEE298 pKa = 4.56VVADD302 pKa = 3.85AGEE305 pKa = 4.0RR306 pKa = 11.84LAWRR310 pKa = 11.84IVDD313 pKa = 3.96QTAAWAGSLDD323 pKa = 4.26LLFTEE328 pKa = 4.78YY329 pKa = 11.11APALMGTLLALRR341 pKa = 11.84TDD343 pKa = 3.81EE344 pKa = 5.24AEE346 pKa = 3.91LTARR350 pKa = 11.84LQSDD354 pKa = 3.59HH355 pKa = 7.17LIGRR359 pKa = 11.84LPAPSSWTDD368 pKa = 2.11WRR370 pKa = 11.84TQHH373 pKa = 7.05GIPNSTTTEE382 pKa = 3.75THH384 pKa = 5.68
Molecular weight: 42.65 kDa
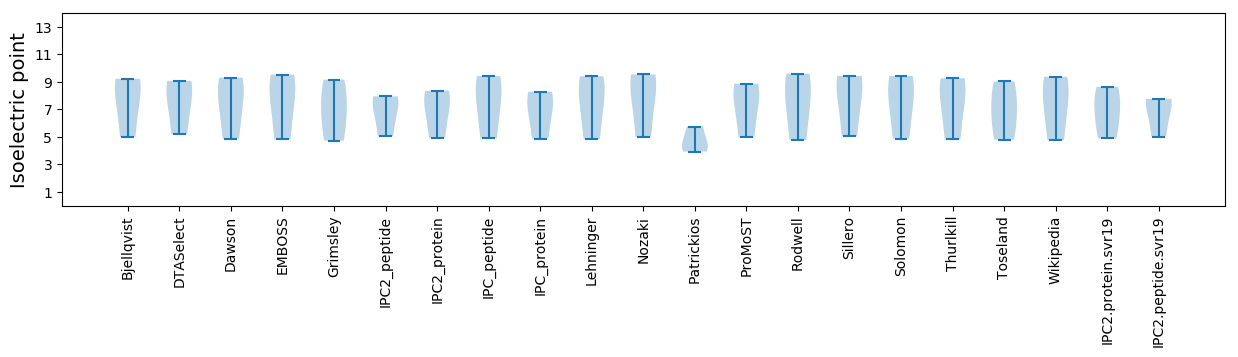
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH35|A0A1L3KH35_9VIRU Uncharacterized protein OS=Wenzhou tombus-like virus 9 OX=1923679 PE=4 SV=1
MM1 pKa = 7.5EE2 pKa = 4.76RR3 pKa = 11.84TLDD6 pKa = 3.84TEE8 pKa = 4.54SLDD11 pKa = 4.01TNSSKK16 pKa = 11.03VGSGCSQVSGLTSDD30 pKa = 5.43DD31 pKa = 3.61GVSCVGSSVASSEE44 pKa = 3.98EE45 pKa = 3.94SRR47 pKa = 11.84TGDD50 pKa = 4.54RR51 pKa = 11.84EE52 pKa = 4.21NRR54 pKa = 11.84WKK56 pKa = 10.87KK57 pKa = 8.39PVRR60 pKa = 11.84RR61 pKa = 11.84TVHH64 pKa = 5.97AVGAAATFGGIVLGCGKK81 pKa = 10.04IGLVLSPVTLGVMWALRR98 pKa = 11.84DD99 pKa = 3.76KK100 pKa = 10.92KK101 pKa = 10.27PSKK104 pKa = 10.5EE105 pKa = 3.7ADD107 pKa = 3.32LVLGVVNGDD116 pKa = 3.29IEE118 pKa = 4.47VEE120 pKa = 4.21KK121 pKa = 10.49EE122 pKa = 3.49ATLASTCEE130 pKa = 4.46GEE132 pKa = 4.12QQPRR136 pKa = 11.84KK137 pKa = 9.59IVGGGRR143 pKa = 11.84NDD145 pKa = 3.63AEE147 pKa = 4.32VVKK150 pKa = 9.73TKK152 pKa = 10.55KK153 pKa = 9.22KK154 pKa = 9.48QRR156 pKa = 11.84WYY158 pKa = 10.42QLKK161 pKa = 9.39KK162 pKa = 8.9QPKK165 pKa = 8.46RR166 pKa = 11.84RR167 pKa = 11.84IPVLAGEE174 pKa = 4.46VASGLKK180 pKa = 9.55VRR182 pKa = 11.84HH183 pKa = 6.0GLLADD188 pKa = 3.43NAEE191 pKa = 3.89NRR193 pKa = 11.84RR194 pKa = 11.84LIRR197 pKa = 11.84SDD199 pKa = 2.65ASRR202 pKa = 11.84RR203 pKa = 11.84CEE205 pKa = 3.62ALRR208 pKa = 11.84RR209 pKa = 11.84DD210 pKa = 3.17GDD212 pKa = 3.53EE213 pKa = 3.62WFKK216 pKa = 11.31NLRR219 pKa = 11.84NHH221 pKa = 7.26DD222 pKa = 3.7MLAVVVHH229 pKa = 6.49ASQMFWLMSDD239 pKa = 4.08DD240 pKa = 3.85EE241 pKa = 4.31EE242 pKa = 5.04AAISIYY248 pKa = 10.43DD249 pKa = 3.61DD250 pKa = 4.04HH251 pKa = 8.64YY252 pKa = 11.53LVANRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84AKK262 pKa = 10.24LASSPTACC270 pKa = 4.99
MM1 pKa = 7.5EE2 pKa = 4.76RR3 pKa = 11.84TLDD6 pKa = 3.84TEE8 pKa = 4.54SLDD11 pKa = 4.01TNSSKK16 pKa = 11.03VGSGCSQVSGLTSDD30 pKa = 5.43DD31 pKa = 3.61GVSCVGSSVASSEE44 pKa = 3.98EE45 pKa = 3.94SRR47 pKa = 11.84TGDD50 pKa = 4.54RR51 pKa = 11.84EE52 pKa = 4.21NRR54 pKa = 11.84WKK56 pKa = 10.87KK57 pKa = 8.39PVRR60 pKa = 11.84RR61 pKa = 11.84TVHH64 pKa = 5.97AVGAAATFGGIVLGCGKK81 pKa = 10.04IGLVLSPVTLGVMWALRR98 pKa = 11.84DD99 pKa = 3.76KK100 pKa = 10.92KK101 pKa = 10.27PSKK104 pKa = 10.5EE105 pKa = 3.7ADD107 pKa = 3.32LVLGVVNGDD116 pKa = 3.29IEE118 pKa = 4.47VEE120 pKa = 4.21KK121 pKa = 10.49EE122 pKa = 3.49ATLASTCEE130 pKa = 4.46GEE132 pKa = 4.12QQPRR136 pKa = 11.84KK137 pKa = 9.59IVGGGRR143 pKa = 11.84NDD145 pKa = 3.63AEE147 pKa = 4.32VVKK150 pKa = 9.73TKK152 pKa = 10.55KK153 pKa = 9.22KK154 pKa = 9.48QRR156 pKa = 11.84WYY158 pKa = 10.42QLKK161 pKa = 9.39KK162 pKa = 8.9QPKK165 pKa = 8.46RR166 pKa = 11.84RR167 pKa = 11.84IPVLAGEE174 pKa = 4.46VASGLKK180 pKa = 9.55VRR182 pKa = 11.84HH183 pKa = 6.0GLLADD188 pKa = 3.43NAEE191 pKa = 3.89NRR193 pKa = 11.84RR194 pKa = 11.84LIRR197 pKa = 11.84SDD199 pKa = 2.65ASRR202 pKa = 11.84RR203 pKa = 11.84CEE205 pKa = 3.62ALRR208 pKa = 11.84RR209 pKa = 11.84DD210 pKa = 3.17GDD212 pKa = 3.53EE213 pKa = 3.62WFKK216 pKa = 11.31NLRR219 pKa = 11.84NHH221 pKa = 7.26DD222 pKa = 3.7MLAVVVHH229 pKa = 6.49ASQMFWLMSDD239 pKa = 4.08DD240 pKa = 3.85EE241 pKa = 4.31EE242 pKa = 5.04AAISIYY248 pKa = 10.43DD249 pKa = 3.61DD250 pKa = 4.04HH251 pKa = 8.64YY252 pKa = 11.53LVANRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84AKK262 pKa = 10.24LASSPTACC270 pKa = 4.99
Molecular weight: 29.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1211 |
270 |
557 |
403.7 |
45.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.193 ± 1.114 | 1.569 ± 0.481 |
6.936 ± 0.075 | 6.276 ± 0.403 |
3.716 ± 0.811 | 7.349 ± 0.497 |
2.395 ± 0.184 | 4.542 ± 0.449 |
5.037 ± 1.069 | 8.175 ± 0.69 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.725 ± 0.397 | 3.633 ± 0.194 |
4.377 ± 0.572 | 2.973 ± 0.351 |
7.845 ± 0.766 | 8.258 ± 1.377 |
5.945 ± 1.486 | 7.184 ± 0.743 |
1.734 ± 0.173 | 3.138 ± 0.613 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
