
Thiosulfativibrio zosterae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Piscirickettsiaceae; Thiosulfativibrio
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
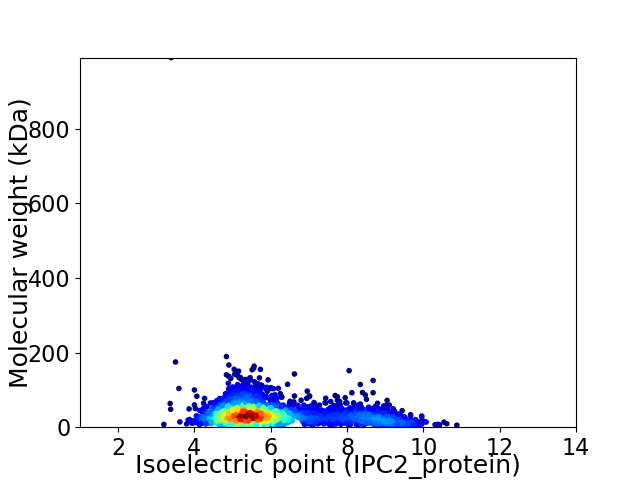
Virtual 2D-PAGE plot for 2362 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6F8PKM1|A0A6F8PKM1_9GAMM tRNA pseudouridine synthase D OS=Thiosulfativibrio zosterae OX=2675053 GN=truD PE=3 SV=1
MM1 pKa = 7.24NCKK4 pKa = 9.77LHH6 pKa = 5.6YY7 pKa = 9.84PKK9 pKa = 10.77VFLISLFIGGLSGCDD24 pKa = 3.19SPSKK28 pKa = 10.71LDD30 pKa = 3.43AEE32 pKa = 5.14GIGKK36 pKa = 9.82YY37 pKa = 10.02ISGVGNIVMSGLVVDD52 pKa = 4.58PAIGGATVYY61 pKa = 10.44IDD63 pKa = 3.7ANGDD67 pKa = 3.67GVIDD71 pKa = 5.14DD72 pKa = 5.51NDD74 pKa = 3.81PQAQTDD80 pKa = 3.6SLGYY84 pKa = 10.2FNYY87 pKa = 10.65NPVTNVDD94 pKa = 3.6YY95 pKa = 11.17CAVGPSAYY103 pKa = 9.49CFPMKK108 pKa = 10.3EE109 pKa = 4.02EE110 pKa = 3.96YY111 pKa = 10.39AGQVLRR117 pKa = 11.84SSGGTDD123 pKa = 3.15LVTNKK128 pKa = 9.65PFEE131 pKa = 4.52GEE133 pKa = 4.08MQLKK137 pKa = 10.35LSSEE141 pKa = 4.16VNEE144 pKa = 4.32NQTEE148 pKa = 4.67GIVITPLSSIVAFMTDD164 pKa = 3.15EE165 pKa = 4.14QKK167 pKa = 11.44ASLKK171 pKa = 10.85SLLNLNEE178 pKa = 4.15SDD180 pKa = 5.43DD181 pKa = 4.25LNKK184 pKa = 10.26LDD186 pKa = 5.28PLDD189 pKa = 4.63FSDD192 pKa = 4.84AKK194 pKa = 11.19YY195 pKa = 8.05PTAEE199 pKa = 4.69DD200 pKa = 3.73KK201 pKa = 11.17AKK203 pKa = 9.92QQQRR207 pKa = 11.84LKK209 pKa = 10.67IIVNIQNTVQALTSALATSDD229 pKa = 3.14KK230 pKa = 9.76TQNNALNEE238 pKa = 3.83AVYY241 pKa = 10.54AVVAQNVTSFTTDD254 pKa = 2.89LQRR257 pKa = 11.84ADD259 pKa = 3.83NQPTNTDD266 pKa = 3.02NSVDD270 pKa = 3.7SLLDD274 pKa = 3.3KK275 pKa = 11.16VIIQVSQSTKK285 pKa = 9.66VSNAVKK291 pKa = 8.79TQAQSLSTTSPNANPTTRR309 pKa = 11.84NALLNNVSALSAASNNVFNTTTITTNANASKK340 pKa = 10.81VLFKK344 pKa = 10.9TEE346 pKa = 4.2VVGEE350 pKa = 4.38TVNATPFDD358 pKa = 3.56ITNTQTAITIINSTVVNNLIDD379 pKa = 4.09GSTADD384 pKa = 4.05FTQLTNIAITQNITSEE400 pKa = 4.29EE401 pKa = 4.07EE402 pKa = 3.6LGGIIDD408 pKa = 4.13SGVGTGDD415 pKa = 3.62PVPGNTDD422 pKa = 2.47IGTTTGDD429 pKa = 3.14GTTTGDD435 pKa = 3.19GTTTGDD441 pKa = 3.19GTTTGDD447 pKa = 3.19GTTTGDD453 pKa = 3.17GTTTGGGGTTIPPITNTAPVAVDD476 pKa = 3.59DD477 pKa = 5.21SITAIEE483 pKa = 4.28DD484 pKa = 3.81TVFTSTIDD492 pKa = 3.7LDD494 pKa = 5.31ANDD497 pKa = 4.75TDD499 pKa = 5.09LDD501 pKa = 4.56SNTLSVVAGTFSTAQGGTLILASNGSYY528 pKa = 10.11TYY530 pKa = 11.14LSATNFNGTDD540 pKa = 3.28TVDD543 pKa = 3.34YY544 pKa = 9.75TVTDD548 pKa = 3.86GALTDD553 pKa = 3.89IGTLTITVEE562 pKa = 4.23PVNDD566 pKa = 3.52LPTANGEE573 pKa = 4.52SITVSKK579 pKa = 10.62DD580 pKa = 3.03VEE582 pKa = 4.69YY583 pKa = 10.62ISSTVNLLANDD594 pKa = 3.94TDD596 pKa = 3.63VDD598 pKa = 4.11GDD600 pKa = 4.0DD601 pKa = 4.12LSIVSGTYY609 pKa = 6.36TTQAGGSITLNANGSFSYY627 pKa = 10.55TPAANFNGTDD637 pKa = 3.13AYY639 pKa = 10.95VYY641 pKa = 10.0EE642 pKa = 4.31VSDD645 pKa = 4.14DD646 pKa = 3.93GGVNASATLTIIVTDD661 pKa = 3.85AMSDD665 pKa = 3.17ADD667 pKa = 4.01EE668 pKa = 4.51TVTTAEE674 pKa = 4.19DD675 pKa = 3.34TALTGNVMSNLTDD688 pKa = 3.97ADD690 pKa = 4.06SSSHH694 pKa = 5.81SVTSFTVEE702 pKa = 3.77GQTYY706 pKa = 9.62QPGSTATITGGTITIAATGAYY727 pKa = 7.86TFTSSEE733 pKa = 4.08NFNGSVPAISYY744 pKa = 11.46AMVDD748 pKa = 3.27NTDD751 pKa = 4.11ANDD754 pKa = 3.81TDD756 pKa = 3.8TSTLNISVTAVTDD769 pKa = 3.58AMSDD773 pKa = 3.22ADD775 pKa = 4.02EE776 pKa = 4.5TLTTAEE782 pKa = 4.27DD783 pKa = 3.5TALTGNVMSNLTDD796 pKa = 3.97ADD798 pKa = 4.06SSSHH802 pKa = 5.81SVTSFTVEE810 pKa = 3.77GQTYY814 pKa = 9.62QPGSTATITGGTITIAATGAYY835 pKa = 7.9TFTPSEE841 pKa = 4.27NFNGTVPAISYY852 pKa = 11.5AMVDD856 pKa = 3.68NNDD859 pKa = 3.32ATDD862 pKa = 3.67TDD864 pKa = 4.0TSTLNISVNSVDD876 pKa = 4.1DD877 pKa = 3.87VPVAVEE883 pKa = 5.59DD884 pKa = 3.87NFTLNEE890 pKa = 4.65DD891 pKa = 3.76GAITDD896 pKa = 3.92TLATNDD902 pKa = 4.06TLSGDD907 pKa = 3.63GGNVFTKK914 pKa = 10.1ATDD917 pKa = 3.81PAHH920 pKa = 5.71GTVTVNADD928 pKa = 3.18GTFTYY933 pKa = 10.34TPAANYY939 pKa = 10.16NGADD943 pKa = 3.52SFSYY947 pKa = 10.46TITDD951 pKa = 3.41ADD953 pKa = 4.36GDD955 pKa = 3.92TSTAVVNLTVNSVDD969 pKa = 3.8DD970 pKa = 3.92VPVAVEE976 pKa = 5.59DD977 pKa = 3.87NFTLNEE983 pKa = 4.65DD984 pKa = 3.76GAITDD989 pKa = 3.92TLATNDD995 pKa = 4.06TLSGDD1000 pKa = 3.63GGNVFTKK1007 pKa = 9.63EE1008 pKa = 3.71TDD1010 pKa = 3.62PAHH1013 pKa = 5.72GTVTVNADD1021 pKa = 3.18GTFTYY1026 pKa = 10.34TPAANYY1032 pKa = 10.16NGADD1036 pKa = 3.52SFSYY1040 pKa = 10.46TITDD1044 pKa = 3.41ADD1046 pKa = 4.36GDD1048 pKa = 3.92TSTAVVNITVVAVTNAMSDD1067 pKa = 3.24ADD1069 pKa = 3.96EE1070 pKa = 4.32TLSTNEE1076 pKa = 3.81DD1077 pKa = 3.38TALSGNVMSNLTDD1090 pKa = 3.97ADD1092 pKa = 4.06SSSHH1096 pKa = 5.78SVTSFSVAGVATSYY1110 pKa = 11.0TPGQTATIVNLGTITIATNGDD1131 pKa = 3.72YY1132 pKa = 10.95TFTPAANFNGSVPAISYY1149 pKa = 11.56AMVDD1153 pKa = 3.47NNDD1156 pKa = 3.84ANDD1159 pKa = 3.91TDD1161 pKa = 3.92TSTLNITVDD1170 pKa = 3.65AVNDD1174 pKa = 3.89SPVALLASTSSPIIGNPFDD1193 pKa = 4.74LEE1195 pKa = 4.33SGLTAEE1201 pKa = 5.72GLIEE1205 pKa = 4.66LDD1207 pKa = 3.04NHH1209 pKa = 6.85KK1210 pKa = 10.49ILIYY1214 pKa = 10.84GLDD1217 pKa = 3.73KK1218 pKa = 11.05INNKK1222 pKa = 10.2LYY1224 pKa = 9.55ISRR1227 pKa = 11.84WFANGRR1233 pKa = 11.84VDD1235 pKa = 3.39TTYY1238 pKa = 9.58GTDD1241 pKa = 4.25GILTVNVDD1249 pKa = 3.55ANAEE1253 pKa = 3.74IDD1255 pKa = 3.85AVKK1258 pKa = 10.73QSDD1261 pKa = 3.7GKK1263 pKa = 11.25VNFLIAYY1270 pKa = 5.91PTAGAAQIKK1279 pKa = 9.9IMQLDD1284 pKa = 3.63SEE1286 pKa = 4.9GVVNITFGTSGVKK1299 pKa = 9.19QLSIINSPSTFTQVRR1314 pKa = 11.84LLMQGTKK1321 pKa = 9.69MIVLGLDD1328 pKa = 3.42DD1329 pKa = 4.3SSNTLVMRR1337 pKa = 11.84LLSTGVYY1344 pKa = 7.99DD1345 pKa = 3.69TTFGDD1350 pKa = 3.83SSNGLAYY1357 pKa = 10.4GEE1359 pKa = 4.62GFLDD1363 pKa = 3.68NLSDD1367 pKa = 3.94VVIGSDD1373 pKa = 3.22DD1374 pKa = 3.61TIYY1377 pKa = 11.63YY1378 pKa = 8.6MGAYY1382 pKa = 9.98NSDD1385 pKa = 2.88IFISKK1390 pKa = 10.29LNADD1394 pKa = 3.8GTINAAFNSNARR1406 pKa = 11.84LEE1408 pKa = 4.62LIIDD1412 pKa = 3.65ALGVFTASNLVLDD1425 pKa = 5.5GSALMYY1431 pKa = 10.88LLAYY1435 pKa = 9.58KK1436 pKa = 10.78DD1437 pKa = 3.55EE1438 pKa = 4.84DD1439 pKa = 4.37SNNKK1443 pKa = 9.88LILGKK1448 pKa = 10.56VSAADD1453 pKa = 3.89GSAVIDD1459 pKa = 3.82SYY1461 pKa = 11.84FSLGYY1466 pKa = 10.3KK1467 pKa = 9.89ILDD1470 pKa = 3.97LSDD1473 pKa = 3.86APQMFPTSVEE1483 pKa = 3.88MLPNNGLLIGGYY1495 pKa = 9.67NPNTVNSISNTLKK1508 pKa = 10.44LAKK1511 pKa = 9.96FNYY1514 pKa = 8.56STGDD1518 pKa = 3.4LEE1520 pKa = 4.89YY1521 pKa = 10.85SFSGNAGVYY1530 pKa = 10.13ISQSVGKK1537 pKa = 10.31VSDD1540 pKa = 3.22IALIGTNNDD1549 pKa = 4.16EE1550 pKa = 3.62IWMLGDD1556 pKa = 3.66RR1557 pKa = 11.84LSEE1560 pKa = 4.21SVGNNGTFNITRR1572 pKa = 11.84LDD1574 pKa = 3.55LGGVVVNYY1582 pKa = 10.74DD1583 pKa = 3.33LAYY1586 pKa = 10.57DD1587 pKa = 3.86VNNGVLPTAVSVSIPAFDD1605 pKa = 3.97VEE1607 pKa = 4.81GDD1609 pKa = 3.33NFTYY1613 pKa = 10.87SLLGTASNAASVTLDD1628 pKa = 3.21SATGEE1633 pKa = 3.93LSYY1636 pKa = 11.84SNDD1639 pKa = 2.72NCYY1642 pKa = 10.56AAGTQVDD1649 pKa = 4.39GLGHH1653 pKa = 6.42YY1654 pKa = 10.18YY1655 pKa = 11.0DD1656 pKa = 5.38LVDD1659 pKa = 5.58FNMTDD1664 pKa = 3.16SQGATSSTQTFTVRR1678 pKa = 11.84LFNCGAA1684 pKa = 3.72
MM1 pKa = 7.24NCKK4 pKa = 9.77LHH6 pKa = 5.6YY7 pKa = 9.84PKK9 pKa = 10.77VFLISLFIGGLSGCDD24 pKa = 3.19SPSKK28 pKa = 10.71LDD30 pKa = 3.43AEE32 pKa = 5.14GIGKK36 pKa = 9.82YY37 pKa = 10.02ISGVGNIVMSGLVVDD52 pKa = 4.58PAIGGATVYY61 pKa = 10.44IDD63 pKa = 3.7ANGDD67 pKa = 3.67GVIDD71 pKa = 5.14DD72 pKa = 5.51NDD74 pKa = 3.81PQAQTDD80 pKa = 3.6SLGYY84 pKa = 10.2FNYY87 pKa = 10.65NPVTNVDD94 pKa = 3.6YY95 pKa = 11.17CAVGPSAYY103 pKa = 9.49CFPMKK108 pKa = 10.3EE109 pKa = 4.02EE110 pKa = 3.96YY111 pKa = 10.39AGQVLRR117 pKa = 11.84SSGGTDD123 pKa = 3.15LVTNKK128 pKa = 9.65PFEE131 pKa = 4.52GEE133 pKa = 4.08MQLKK137 pKa = 10.35LSSEE141 pKa = 4.16VNEE144 pKa = 4.32NQTEE148 pKa = 4.67GIVITPLSSIVAFMTDD164 pKa = 3.15EE165 pKa = 4.14QKK167 pKa = 11.44ASLKK171 pKa = 10.85SLLNLNEE178 pKa = 4.15SDD180 pKa = 5.43DD181 pKa = 4.25LNKK184 pKa = 10.26LDD186 pKa = 5.28PLDD189 pKa = 4.63FSDD192 pKa = 4.84AKK194 pKa = 11.19YY195 pKa = 8.05PTAEE199 pKa = 4.69DD200 pKa = 3.73KK201 pKa = 11.17AKK203 pKa = 9.92QQQRR207 pKa = 11.84LKK209 pKa = 10.67IIVNIQNTVQALTSALATSDD229 pKa = 3.14KK230 pKa = 9.76TQNNALNEE238 pKa = 3.83AVYY241 pKa = 10.54AVVAQNVTSFTTDD254 pKa = 2.89LQRR257 pKa = 11.84ADD259 pKa = 3.83NQPTNTDD266 pKa = 3.02NSVDD270 pKa = 3.7SLLDD274 pKa = 3.3KK275 pKa = 11.16VIIQVSQSTKK285 pKa = 9.66VSNAVKK291 pKa = 8.79TQAQSLSTTSPNANPTTRR309 pKa = 11.84NALLNNVSALSAASNNVFNTTTITTNANASKK340 pKa = 10.81VLFKK344 pKa = 10.9TEE346 pKa = 4.2VVGEE350 pKa = 4.38TVNATPFDD358 pKa = 3.56ITNTQTAITIINSTVVNNLIDD379 pKa = 4.09GSTADD384 pKa = 4.05FTQLTNIAITQNITSEE400 pKa = 4.29EE401 pKa = 4.07EE402 pKa = 3.6LGGIIDD408 pKa = 4.13SGVGTGDD415 pKa = 3.62PVPGNTDD422 pKa = 2.47IGTTTGDD429 pKa = 3.14GTTTGDD435 pKa = 3.19GTTTGDD441 pKa = 3.19GTTTGDD447 pKa = 3.19GTTTGDD453 pKa = 3.17GTTTGGGGTTIPPITNTAPVAVDD476 pKa = 3.59DD477 pKa = 5.21SITAIEE483 pKa = 4.28DD484 pKa = 3.81TVFTSTIDD492 pKa = 3.7LDD494 pKa = 5.31ANDD497 pKa = 4.75TDD499 pKa = 5.09LDD501 pKa = 4.56SNTLSVVAGTFSTAQGGTLILASNGSYY528 pKa = 10.11TYY530 pKa = 11.14LSATNFNGTDD540 pKa = 3.28TVDD543 pKa = 3.34YY544 pKa = 9.75TVTDD548 pKa = 3.86GALTDD553 pKa = 3.89IGTLTITVEE562 pKa = 4.23PVNDD566 pKa = 3.52LPTANGEE573 pKa = 4.52SITVSKK579 pKa = 10.62DD580 pKa = 3.03VEE582 pKa = 4.69YY583 pKa = 10.62ISSTVNLLANDD594 pKa = 3.94TDD596 pKa = 3.63VDD598 pKa = 4.11GDD600 pKa = 4.0DD601 pKa = 4.12LSIVSGTYY609 pKa = 6.36TTQAGGSITLNANGSFSYY627 pKa = 10.55TPAANFNGTDD637 pKa = 3.13AYY639 pKa = 10.95VYY641 pKa = 10.0EE642 pKa = 4.31VSDD645 pKa = 4.14DD646 pKa = 3.93GGVNASATLTIIVTDD661 pKa = 3.85AMSDD665 pKa = 3.17ADD667 pKa = 4.01EE668 pKa = 4.51TVTTAEE674 pKa = 4.19DD675 pKa = 3.34TALTGNVMSNLTDD688 pKa = 3.97ADD690 pKa = 4.06SSSHH694 pKa = 5.81SVTSFTVEE702 pKa = 3.77GQTYY706 pKa = 9.62QPGSTATITGGTITIAATGAYY727 pKa = 7.86TFTSSEE733 pKa = 4.08NFNGSVPAISYY744 pKa = 11.46AMVDD748 pKa = 3.27NTDD751 pKa = 4.11ANDD754 pKa = 3.81TDD756 pKa = 3.8TSTLNISVTAVTDD769 pKa = 3.58AMSDD773 pKa = 3.22ADD775 pKa = 4.02EE776 pKa = 4.5TLTTAEE782 pKa = 4.27DD783 pKa = 3.5TALTGNVMSNLTDD796 pKa = 3.97ADD798 pKa = 4.06SSSHH802 pKa = 5.81SVTSFTVEE810 pKa = 3.77GQTYY814 pKa = 9.62QPGSTATITGGTITIAATGAYY835 pKa = 7.9TFTPSEE841 pKa = 4.27NFNGTVPAISYY852 pKa = 11.5AMVDD856 pKa = 3.68NNDD859 pKa = 3.32ATDD862 pKa = 3.67TDD864 pKa = 4.0TSTLNISVNSVDD876 pKa = 4.1DD877 pKa = 3.87VPVAVEE883 pKa = 5.59DD884 pKa = 3.87NFTLNEE890 pKa = 4.65DD891 pKa = 3.76GAITDD896 pKa = 3.92TLATNDD902 pKa = 4.06TLSGDD907 pKa = 3.63GGNVFTKK914 pKa = 10.1ATDD917 pKa = 3.81PAHH920 pKa = 5.71GTVTVNADD928 pKa = 3.18GTFTYY933 pKa = 10.34TPAANYY939 pKa = 10.16NGADD943 pKa = 3.52SFSYY947 pKa = 10.46TITDD951 pKa = 3.41ADD953 pKa = 4.36GDD955 pKa = 3.92TSTAVVNLTVNSVDD969 pKa = 3.8DD970 pKa = 3.92VPVAVEE976 pKa = 5.59DD977 pKa = 3.87NFTLNEE983 pKa = 4.65DD984 pKa = 3.76GAITDD989 pKa = 3.92TLATNDD995 pKa = 4.06TLSGDD1000 pKa = 3.63GGNVFTKK1007 pKa = 9.63EE1008 pKa = 3.71TDD1010 pKa = 3.62PAHH1013 pKa = 5.72GTVTVNADD1021 pKa = 3.18GTFTYY1026 pKa = 10.34TPAANYY1032 pKa = 10.16NGADD1036 pKa = 3.52SFSYY1040 pKa = 10.46TITDD1044 pKa = 3.41ADD1046 pKa = 4.36GDD1048 pKa = 3.92TSTAVVNITVVAVTNAMSDD1067 pKa = 3.24ADD1069 pKa = 3.96EE1070 pKa = 4.32TLSTNEE1076 pKa = 3.81DD1077 pKa = 3.38TALSGNVMSNLTDD1090 pKa = 3.97ADD1092 pKa = 4.06SSSHH1096 pKa = 5.78SVTSFSVAGVATSYY1110 pKa = 11.0TPGQTATIVNLGTITIATNGDD1131 pKa = 3.72YY1132 pKa = 10.95TFTPAANFNGSVPAISYY1149 pKa = 11.56AMVDD1153 pKa = 3.47NNDD1156 pKa = 3.84ANDD1159 pKa = 3.91TDD1161 pKa = 3.92TSTLNITVDD1170 pKa = 3.65AVNDD1174 pKa = 3.89SPVALLASTSSPIIGNPFDD1193 pKa = 4.74LEE1195 pKa = 4.33SGLTAEE1201 pKa = 5.72GLIEE1205 pKa = 4.66LDD1207 pKa = 3.04NHH1209 pKa = 6.85KK1210 pKa = 10.49ILIYY1214 pKa = 10.84GLDD1217 pKa = 3.73KK1218 pKa = 11.05INNKK1222 pKa = 10.2LYY1224 pKa = 9.55ISRR1227 pKa = 11.84WFANGRR1233 pKa = 11.84VDD1235 pKa = 3.39TTYY1238 pKa = 9.58GTDD1241 pKa = 4.25GILTVNVDD1249 pKa = 3.55ANAEE1253 pKa = 3.74IDD1255 pKa = 3.85AVKK1258 pKa = 10.73QSDD1261 pKa = 3.7GKK1263 pKa = 11.25VNFLIAYY1270 pKa = 5.91PTAGAAQIKK1279 pKa = 9.9IMQLDD1284 pKa = 3.63SEE1286 pKa = 4.9GVVNITFGTSGVKK1299 pKa = 9.19QLSIINSPSTFTQVRR1314 pKa = 11.84LLMQGTKK1321 pKa = 9.69MIVLGLDD1328 pKa = 3.42DD1329 pKa = 4.3SSNTLVMRR1337 pKa = 11.84LLSTGVYY1344 pKa = 7.99DD1345 pKa = 3.69TTFGDD1350 pKa = 3.83SSNGLAYY1357 pKa = 10.4GEE1359 pKa = 4.62GFLDD1363 pKa = 3.68NLSDD1367 pKa = 3.94VVIGSDD1373 pKa = 3.22DD1374 pKa = 3.61TIYY1377 pKa = 11.63YY1378 pKa = 8.6MGAYY1382 pKa = 9.98NSDD1385 pKa = 2.88IFISKK1390 pKa = 10.29LNADD1394 pKa = 3.8GTINAAFNSNARR1406 pKa = 11.84LEE1408 pKa = 4.62LIIDD1412 pKa = 3.65ALGVFTASNLVLDD1425 pKa = 5.5GSALMYY1431 pKa = 10.88LLAYY1435 pKa = 9.58KK1436 pKa = 10.78DD1437 pKa = 3.55EE1438 pKa = 4.84DD1439 pKa = 4.37SNNKK1443 pKa = 9.88LILGKK1448 pKa = 10.56VSAADD1453 pKa = 3.89GSAVIDD1459 pKa = 3.82SYY1461 pKa = 11.84FSLGYY1466 pKa = 10.3KK1467 pKa = 9.89ILDD1470 pKa = 3.97LSDD1473 pKa = 3.86APQMFPTSVEE1483 pKa = 3.88MLPNNGLLIGGYY1495 pKa = 9.67NPNTVNSISNTLKK1508 pKa = 10.44LAKK1511 pKa = 9.96FNYY1514 pKa = 8.56STGDD1518 pKa = 3.4LEE1520 pKa = 4.89YY1521 pKa = 10.85SFSGNAGVYY1530 pKa = 10.13ISQSVGKK1537 pKa = 10.31VSDD1540 pKa = 3.22IALIGTNNDD1549 pKa = 4.16EE1550 pKa = 3.62IWMLGDD1556 pKa = 3.66RR1557 pKa = 11.84LSEE1560 pKa = 4.21SVGNNGTFNITRR1572 pKa = 11.84LDD1574 pKa = 3.55LGGVVVNYY1582 pKa = 10.74DD1583 pKa = 3.33LAYY1586 pKa = 10.57DD1587 pKa = 3.86VNNGVLPTAVSVSIPAFDD1605 pKa = 3.97VEE1607 pKa = 4.81GDD1609 pKa = 3.33NFTYY1613 pKa = 10.87SLLGTASNAASVTLDD1628 pKa = 3.21SATGEE1633 pKa = 3.93LSYY1636 pKa = 11.84SNDD1639 pKa = 2.72NCYY1642 pKa = 10.56AAGTQVDD1649 pKa = 4.39GLGHH1653 pKa = 6.42YY1654 pKa = 10.18YY1655 pKa = 11.0DD1656 pKa = 5.38LVDD1659 pKa = 5.58FNMTDD1664 pKa = 3.16SQGATSSTQTFTVRR1678 pKa = 11.84LFNCGAA1684 pKa = 3.72
Molecular weight: 174.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6F8PRG8|A0A6F8PRG8_9GAMM Probable membrane transporter protein OS=Thiosulfativibrio zosterae OX=2675053 GN=THMIRHAT_23730 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.76GRR39 pKa = 11.84KK40 pKa = 8.93RR41 pKa = 11.84LTVSDD46 pKa = 3.89KK47 pKa = 11.45
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.76GRR39 pKa = 11.84KK40 pKa = 8.93RR41 pKa = 11.84LTVSDD46 pKa = 3.89KK47 pKa = 11.45
Molecular weight: 5.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
802121 |
45 |
9540 |
339.6 |
37.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.946 ± 0.053 | 0.786 ± 0.018 |
5.333 ± 0.092 | 6.214 ± 0.05 |
4.195 ± 0.037 | 6.475 ± 0.051 |
2.248 ± 0.035 | 6.305 ± 0.043 |
5.724 ± 0.047 | 10.844 ± 0.109 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.772 ± 0.037 | 4.35 ± 0.044 |
4.044 ± 0.039 | 5.371 ± 0.058 |
3.953 ± 0.055 | 6.132 ± 0.047 |
5.47 ± 0.144 | 6.729 ± 0.045 |
1.23 ± 0.023 | 2.879 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
