
Colletotrichum fioriniae PJ7
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Glomerellales; Glomerellaceae; Colletotrichum; Colletotrichum acutatum species complex; Colletotrichum fioriniae
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
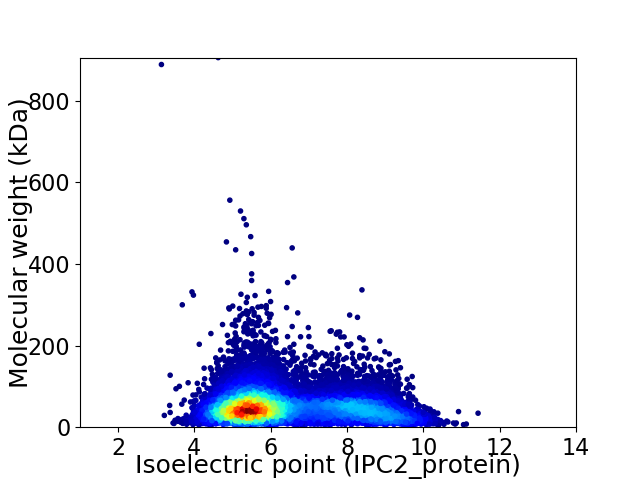
Virtual 2D-PAGE plot for 13759 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A010RL95|A0A010RL95_9PEZI U6 snRNA-associated Sm-like protein LSm3 OS=Colletotrichum fioriniae PJ7 OX=1445577 GN=LSM3 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 10.3LDD4 pKa = 4.17AEE6 pKa = 4.83LFYY9 pKa = 11.16QEE11 pKa = 4.04NWADD15 pKa = 3.49STYY18 pKa = 11.01TLSNWGDD25 pKa = 3.38STIWEE30 pKa = 4.4WGTKK34 pKa = 9.38AVANVANLSTDD45 pKa = 2.67WSNLTDD51 pKa = 4.55GLAEE55 pKa = 4.35DD56 pKa = 4.89TPHH59 pKa = 7.58GDD61 pKa = 3.05ACADD65 pKa = 3.67NIARR69 pKa = 11.84DD70 pKa = 4.01DD71 pKa = 3.9NGVCVTGDD79 pKa = 3.37ASTDD83 pKa = 2.72ICAGFDD89 pKa = 3.48TFSVHH94 pKa = 6.97SLDD97 pKa = 3.97RR98 pKa = 11.84VSTEE102 pKa = 4.83DD103 pKa = 3.27IADD106 pKa = 3.41VGNFILVNSSTEE118 pKa = 3.94DD119 pKa = 3.26LSAEE123 pKa = 4.17NGGSVTPCHH132 pKa = 6.37
MM1 pKa = 7.45KK2 pKa = 10.3LDD4 pKa = 4.17AEE6 pKa = 4.83LFYY9 pKa = 11.16QEE11 pKa = 4.04NWADD15 pKa = 3.49STYY18 pKa = 11.01TLSNWGDD25 pKa = 3.38STIWEE30 pKa = 4.4WGTKK34 pKa = 9.38AVANVANLSTDD45 pKa = 2.67WSNLTDD51 pKa = 4.55GLAEE55 pKa = 4.35DD56 pKa = 4.89TPHH59 pKa = 7.58GDD61 pKa = 3.05ACADD65 pKa = 3.67NIARR69 pKa = 11.84DD70 pKa = 4.01DD71 pKa = 3.9NGVCVTGDD79 pKa = 3.37ASTDD83 pKa = 2.72ICAGFDD89 pKa = 3.48TFSVHH94 pKa = 6.97SLDD97 pKa = 3.97RR98 pKa = 11.84VSTEE102 pKa = 4.83DD103 pKa = 3.27IADD106 pKa = 3.41VGNFILVNSSTEE118 pKa = 3.94DD119 pKa = 3.26LSAEE123 pKa = 4.17NGGSVTPCHH132 pKa = 6.37
Molecular weight: 14.13 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A010RBN8|A0A010RBN8_9PEZI Protein kinase OS=Colletotrichum fioriniae PJ7 OX=1445577 GN=CFIO01_10155 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 9.82PLRR5 pKa = 11.84LTTSRR10 pKa = 11.84PVSPTTVVRR19 pKa = 11.84RR20 pKa = 11.84PRR22 pKa = 11.84RR23 pKa = 11.84LRR25 pKa = 11.84PPSVRR30 pKa = 11.84SLPSTRR36 pKa = 11.84LSRR39 pKa = 11.84LVTLRR44 pKa = 11.84LVSPTTAARR53 pKa = 11.84RR54 pKa = 11.84PRR56 pKa = 11.84RR57 pKa = 11.84PRR59 pKa = 11.84PPSVRR64 pKa = 11.84SLLSMRR70 pKa = 11.84PSRR73 pKa = 11.84PATLSLASLTTAARR87 pKa = 11.84RR88 pKa = 11.84PRR90 pKa = 11.84RR91 pKa = 11.84PRR93 pKa = 11.84PPSATNPPSTRR104 pKa = 11.84PSRR107 pKa = 11.84LVTSKK112 pKa = 10.16PASPTTAARR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84PRR127 pKa = 11.84LPSVTNSSPRR137 pKa = 11.84RR138 pKa = 11.84PSSLLSTRR146 pKa = 11.84SRR148 pKa = 11.84PVSPTTAARR157 pKa = 11.84RR158 pKa = 11.84PRR160 pKa = 11.84RR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84PPSAMSPPPTTPSSPPSTRR182 pKa = 11.84SRR184 pKa = 11.84PASLTTVARR193 pKa = 11.84RR194 pKa = 11.84PRR196 pKa = 11.84RR197 pKa = 11.84QRR199 pKa = 11.84LLSARR204 pKa = 11.84SPPPTTPLSTPSRR217 pKa = 11.84PASPTTVXXXRR228 pKa = 11.84RR229 pKa = 11.84PRR231 pKa = 11.84PRR233 pKa = 11.84SATSSLPRR241 pKa = 11.84TLPSTPLRR249 pKa = 11.84LVSPTTAARR258 pKa = 11.84RR259 pKa = 11.84PRR261 pKa = 11.84RR262 pKa = 11.84PRR264 pKa = 11.84PRR266 pKa = 11.84SATSTPPLRR275 pKa = 11.84LALSPLKK282 pKa = 10.79GVVQLCGRR290 pKa = 11.84HH291 pKa = 6.26GGQQLNALTRR301 pKa = 11.84ATLEE305 pKa = 3.79LRR307 pKa = 11.84RR308 pKa = 11.84PPVLPCII315 pKa = 4.39
MM1 pKa = 7.51KK2 pKa = 9.82PLRR5 pKa = 11.84LTTSRR10 pKa = 11.84PVSPTTVVRR19 pKa = 11.84RR20 pKa = 11.84PRR22 pKa = 11.84RR23 pKa = 11.84LRR25 pKa = 11.84PPSVRR30 pKa = 11.84SLPSTRR36 pKa = 11.84LSRR39 pKa = 11.84LVTLRR44 pKa = 11.84LVSPTTAARR53 pKa = 11.84RR54 pKa = 11.84PRR56 pKa = 11.84RR57 pKa = 11.84PRR59 pKa = 11.84PPSVRR64 pKa = 11.84SLLSMRR70 pKa = 11.84PSRR73 pKa = 11.84PATLSLASLTTAARR87 pKa = 11.84RR88 pKa = 11.84PRR90 pKa = 11.84RR91 pKa = 11.84PRR93 pKa = 11.84PPSATNPPSTRR104 pKa = 11.84PSRR107 pKa = 11.84LVTSKK112 pKa = 10.16PASPTTAARR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84PRR127 pKa = 11.84LPSVTNSSPRR137 pKa = 11.84RR138 pKa = 11.84PSSLLSTRR146 pKa = 11.84SRR148 pKa = 11.84PVSPTTAARR157 pKa = 11.84RR158 pKa = 11.84PRR160 pKa = 11.84RR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84PPSAMSPPPTTPSSPPSTRR182 pKa = 11.84SRR184 pKa = 11.84PASLTTVARR193 pKa = 11.84RR194 pKa = 11.84PRR196 pKa = 11.84RR197 pKa = 11.84QRR199 pKa = 11.84LLSARR204 pKa = 11.84SPPPTTPLSTPSRR217 pKa = 11.84PASPTTVXXXRR228 pKa = 11.84RR229 pKa = 11.84PRR231 pKa = 11.84PRR233 pKa = 11.84SATSSLPRR241 pKa = 11.84TLPSTPLRR249 pKa = 11.84LVSPTTAARR258 pKa = 11.84RR259 pKa = 11.84PRR261 pKa = 11.84RR262 pKa = 11.84PRR264 pKa = 11.84PRR266 pKa = 11.84SATSTPPLRR275 pKa = 11.84LALSPLKK282 pKa = 10.79GVVQLCGRR290 pKa = 11.84HH291 pKa = 6.26GGQQLNALTRR301 pKa = 11.84ATLEE305 pKa = 3.79LRR307 pKa = 11.84RR308 pKa = 11.84PPVLPCII315 pKa = 4.39
Molecular weight: 34.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6816730 |
24 |
9111 |
495.4 |
54.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.073 ± 0.019 | 1.217 ± 0.007 |
5.714 ± 0.014 | 6.03 ± 0.023 |
3.79 ± 0.012 | 7.152 ± 0.02 |
2.312 ± 0.011 | 4.795 ± 0.014 |
4.804 ± 0.017 | 8.678 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.21 ± 0.008 | 3.672 ± 0.01 |
6.102 ± 0.026 | 3.889 ± 0.014 |
5.864 ± 0.02 | 8.067 ± 0.025 |
6.11 ± 0.025 | 6.255 ± 0.018 |
1.538 ± 0.008 | 2.725 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
