
Rhesus monkey rhadinovirus H26-95
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Gammaherpesvirinae; Rhadinovirus; Macacine gammaherpesvirus 5
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
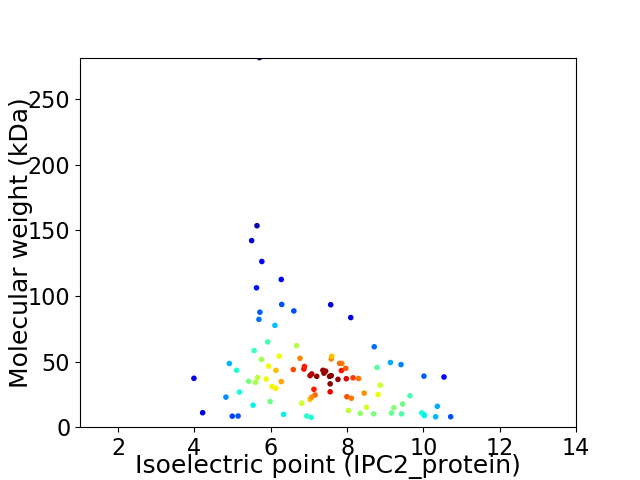
Virtual 2D-PAGE plot for 95 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q77NI9|Q77NI9_9GAMA ORF52 OS=Rhesus monkey rhadinovirus H26-95 OX=69256 GN=ORF52 PE=3 SV=1
MM1 pKa = 7.27TGSIVLALALLACLYY16 pKa = 10.8LCLPVCATVTTSSTTGTGTPPVTTNPSAAPSVTPSFYY53 pKa = 10.66DD54 pKa = 3.56YY55 pKa = 11.29DD56 pKa = 4.35CSADD60 pKa = 3.26TYY62 pKa = 11.31QPVLSSFSSIWAVINSVLVAVATFLYY88 pKa = 8.44LTYY91 pKa = 9.62MCFFKK96 pKa = 10.81FVEE99 pKa = 5.05TVAHH103 pKa = 5.99EE104 pKa = 4.0
MM1 pKa = 7.27TGSIVLALALLACLYY16 pKa = 10.8LCLPVCATVTTSSTTGTGTPPVTTNPSAAPSVTPSFYY53 pKa = 10.66DD54 pKa = 3.56YY55 pKa = 11.29DD56 pKa = 4.35CSADD60 pKa = 3.26TYY62 pKa = 11.31QPVLSSFSSIWAVINSVLVAVATFLYY88 pKa = 8.44LTYY91 pKa = 9.62MCFFKK96 pKa = 10.81FVEE99 pKa = 5.05TVAHH103 pKa = 5.99EE104 pKa = 4.0
Molecular weight: 11.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9J2H1|Q9J2H1_9GAMA Cyclin D homolog OS=Rhesus monkey rhadinovirus H26-95 OX=69256 GN=ORF72 PE=3 SV=1
MM1 pKa = 7.15WGSRR5 pKa = 11.84QHH7 pKa = 6.75RR8 pKa = 11.84SGIVSGHH15 pKa = 5.78GLRR18 pKa = 11.84SSCRR22 pKa = 11.84GHH24 pKa = 6.48CGRR27 pKa = 11.84RR28 pKa = 11.84GGTRR32 pKa = 11.84EE33 pKa = 3.5QAGRR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84GRR42 pKa = 11.84LGRR45 pKa = 11.84ASLPAPTPPAPTTSGPQVRR64 pKa = 11.84AVAEE68 pKa = 4.08QGHH71 pKa = 6.07GSDD74 pKa = 4.09TEE76 pKa = 4.21TATEE80 pKa = 4.15SRR82 pKa = 11.84HH83 pKa = 5.67GSSQGSPSGSGSEE96 pKa = 4.18SVIVLGSPTPSPSGSAPVLASGLSPRR122 pKa = 11.84NTSGSSPASPASHH135 pKa = 6.74SPPPSPPSHH144 pKa = 7.34PGPHH148 pKa = 6.08SPAPPSSHH156 pKa = 6.3NPSPNQQPSSFLQPSPHH173 pKa = 7.33DD174 pKa = 3.77SPEE177 pKa = 4.13PPEE180 pKa = 4.97PPTSLPPPDD189 pKa = 4.45SPGPPQSPTPTSSPPPQSPPDD210 pKa = 3.79SPEE213 pKa = 4.23PPQSPTPQQAPSPNTQQAVSHH234 pKa = 6.71TDD236 pKa = 2.93HH237 pKa = 7.02PTGPSRR243 pKa = 11.84PGPPFPGHH251 pKa = 6.04TSHH254 pKa = 7.1SYY256 pKa = 9.39TVGGWGPPTRR266 pKa = 11.84AGGVPCLRR274 pKa = 11.84LRR276 pKa = 11.84CTSHH280 pKa = 6.87NSHH283 pKa = 6.7EE284 pKa = 4.5DD285 pKa = 3.42EE286 pKa = 4.63APEE289 pKa = 3.91RR290 pKa = 11.84QQEE293 pKa = 4.17QEE295 pKa = 3.97GEE297 pKa = 4.1EE298 pKa = 4.25RR299 pKa = 11.84QQQPARR305 pKa = 11.84PPRR308 pKa = 11.84PPRR311 pKa = 11.84PPRR314 pKa = 11.84YY315 pKa = 8.48PIPIPYY321 pKa = 9.16PSSEE325 pKa = 4.01EE326 pKa = 3.88EE327 pKa = 4.06VPRR330 pKa = 11.84KK331 pKa = 9.69YY332 pKa = 10.3RR333 pKa = 11.84PQRR336 pKa = 11.84RR337 pKa = 11.84FYY339 pKa = 10.68RR340 pKa = 11.84QVLGPRR346 pKa = 11.84IDD348 pKa = 3.73PPRR351 pKa = 11.84PGPWCHH357 pKa = 6.42GVIFCNSDD365 pKa = 3.2PYY367 pKa = 11.37SLYY370 pKa = 10.78RR371 pKa = 11.84LARR374 pKa = 11.84CLQFPGIRR382 pKa = 11.84ASSVRR387 pKa = 11.84VLPDD391 pKa = 3.26APGSPVIPAFCITVFCQSRR410 pKa = 11.84GTAKK414 pKa = 10.28AVKK417 pKa = 9.25KK418 pKa = 10.48ARR420 pKa = 11.84RR421 pKa = 11.84RR422 pKa = 11.84WEE424 pKa = 3.82RR425 pKa = 11.84HH426 pKa = 5.73HH427 pKa = 7.05PSAPHH432 pKa = 5.35FQASIVRR439 pKa = 11.84MDD441 pKa = 3.6RR442 pKa = 11.84GLPIQHH448 pKa = 6.95
MM1 pKa = 7.15WGSRR5 pKa = 11.84QHH7 pKa = 6.75RR8 pKa = 11.84SGIVSGHH15 pKa = 5.78GLRR18 pKa = 11.84SSCRR22 pKa = 11.84GHH24 pKa = 6.48CGRR27 pKa = 11.84RR28 pKa = 11.84GGTRR32 pKa = 11.84EE33 pKa = 3.5QAGRR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84GRR42 pKa = 11.84LGRR45 pKa = 11.84ASLPAPTPPAPTTSGPQVRR64 pKa = 11.84AVAEE68 pKa = 4.08QGHH71 pKa = 6.07GSDD74 pKa = 4.09TEE76 pKa = 4.21TATEE80 pKa = 4.15SRR82 pKa = 11.84HH83 pKa = 5.67GSSQGSPSGSGSEE96 pKa = 4.18SVIVLGSPTPSPSGSAPVLASGLSPRR122 pKa = 11.84NTSGSSPASPASHH135 pKa = 6.74SPPPSPPSHH144 pKa = 7.34PGPHH148 pKa = 6.08SPAPPSSHH156 pKa = 6.3NPSPNQQPSSFLQPSPHH173 pKa = 7.33DD174 pKa = 3.77SPEE177 pKa = 4.13PPEE180 pKa = 4.97PPTSLPPPDD189 pKa = 4.45SPGPPQSPTPTSSPPPQSPPDD210 pKa = 3.79SPEE213 pKa = 4.23PPQSPTPQQAPSPNTQQAVSHH234 pKa = 6.71TDD236 pKa = 2.93HH237 pKa = 7.02PTGPSRR243 pKa = 11.84PGPPFPGHH251 pKa = 6.04TSHH254 pKa = 7.1SYY256 pKa = 9.39TVGGWGPPTRR266 pKa = 11.84AGGVPCLRR274 pKa = 11.84LRR276 pKa = 11.84CTSHH280 pKa = 6.87NSHH283 pKa = 6.7EE284 pKa = 4.5DD285 pKa = 3.42EE286 pKa = 4.63APEE289 pKa = 3.91RR290 pKa = 11.84QQEE293 pKa = 4.17QEE295 pKa = 3.97GEE297 pKa = 4.1EE298 pKa = 4.25RR299 pKa = 11.84QQQPARR305 pKa = 11.84PPRR308 pKa = 11.84PPRR311 pKa = 11.84PPRR314 pKa = 11.84YY315 pKa = 8.48PIPIPYY321 pKa = 9.16PSSEE325 pKa = 4.01EE326 pKa = 3.88EE327 pKa = 4.06VPRR330 pKa = 11.84KK331 pKa = 9.69YY332 pKa = 10.3RR333 pKa = 11.84PQRR336 pKa = 11.84RR337 pKa = 11.84FYY339 pKa = 10.68RR340 pKa = 11.84QVLGPRR346 pKa = 11.84IDD348 pKa = 3.73PPRR351 pKa = 11.84PGPWCHH357 pKa = 6.42GVIFCNSDD365 pKa = 3.2PYY367 pKa = 11.37SLYY370 pKa = 10.78RR371 pKa = 11.84LARR374 pKa = 11.84CLQFPGIRR382 pKa = 11.84ASSVRR387 pKa = 11.84VLPDD391 pKa = 3.26APGSPVIPAFCITVFCQSRR410 pKa = 11.84GTAKK414 pKa = 10.28AVKK417 pKa = 9.25KK418 pKa = 10.48ARR420 pKa = 11.84RR421 pKa = 11.84RR422 pKa = 11.84WEE424 pKa = 3.82RR425 pKa = 11.84HH426 pKa = 5.73HH427 pKa = 7.05PSAPHH432 pKa = 5.35FQASIVRR439 pKa = 11.84MDD441 pKa = 3.6RR442 pKa = 11.84GLPIQHH448 pKa = 6.95
Molecular weight: 47.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
37272 |
69 |
2548 |
392.3 |
43.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.722 ± 0.188 | 2.578 ± 0.167 |
4.668 ± 0.16 | 4.934 ± 0.173 |
4.373 ± 0.158 | 6.359 ± 0.362 |
2.715 ± 0.09 | 4.832 ± 0.18 |
4.014 ± 0.187 | 9.943 ± 0.239 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.066 ± 0.106 | 4.116 ± 0.16 |
6.45 ± 0.278 | 3.689 ± 0.164 |
6.08 ± 0.208 | 7.268 ± 0.182 |
6.581 ± 0.284 | 7.247 ± 0.219 |
1.256 ± 0.099 | 3.11 ± 0.136 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
