
Natronobacterium gregoryi (strain ATCC 43098 / DSM 3393 / CCM 3738 / CIP 104747 / IAM 13177 / JCM 8860 / NBRC 102187 / NCIMB 2189 / SP2)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Natronobacterium; Natronobacterium gregoryi
Average proteome isoelectric point is 4.81
Get precalculated fractions of proteins
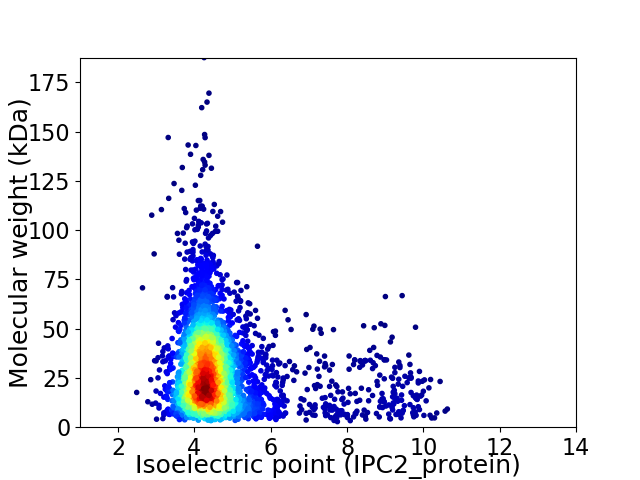
Virtual 2D-PAGE plot for 3624 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
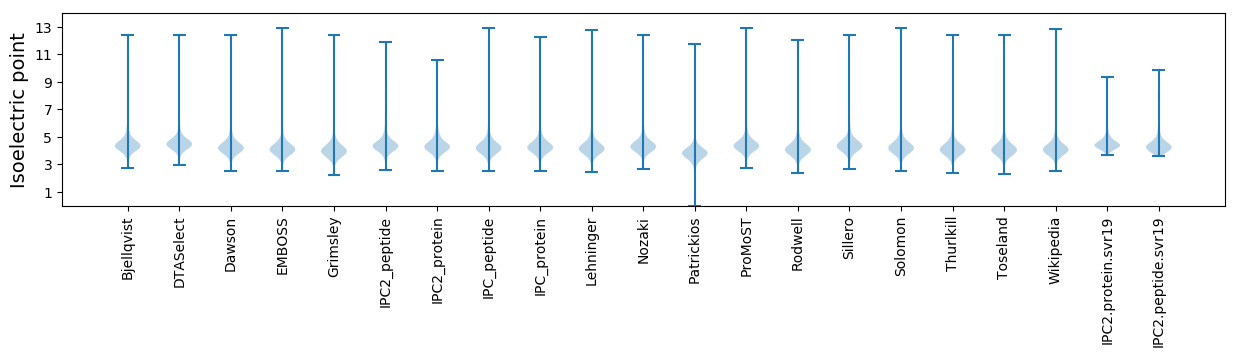
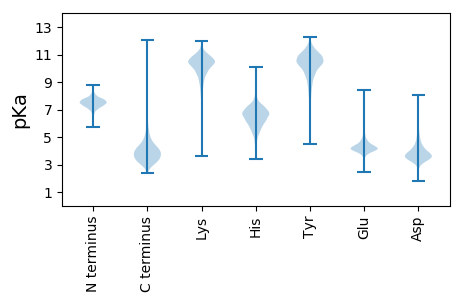
Protein with the lowest isoelectric point:
>tr|L0AKB0|L0AKB0_NATGS ABC-type polar amino acid transport system ATPase component OS=Natronobacterium gregoryi (strain ATCC 43098 / DSM 3393 / CCM 3738 / CIP 104747 / IAM 13177 / JCM 8860 / NBRC 102187 / NCIMB 2189 / SP2) OX=797304 GN=Natgr_3203 PE=4 SV=1
MM1 pKa = 7.85ADD3 pKa = 4.17ALDD6 pKa = 4.18DD7 pKa = 5.97DD8 pKa = 4.6IYY10 pKa = 11.48QRR12 pKa = 11.84TKK14 pKa = 11.12ALLEE18 pKa = 4.39PGEE21 pKa = 4.25IKK23 pKa = 11.07LNGAVVHH30 pKa = 5.86TEE32 pKa = 3.62YY33 pKa = 11.18DD34 pKa = 3.22GGDD37 pKa = 3.85DD38 pKa = 3.74VKK40 pKa = 11.01MMQATIDD47 pKa = 3.66VGDD50 pKa = 4.23VIAAHH55 pKa = 6.52SGHH58 pKa = 7.2DD59 pKa = 3.65PNDD62 pKa = 3.68CYY64 pKa = 11.34VYY66 pKa = 10.73SGNDD70 pKa = 3.42DD71 pKa = 4.02PDD73 pKa = 3.96FSSNQHH79 pKa = 5.89QGLTLDD85 pKa = 4.21DD86 pKa = 4.98EE87 pKa = 4.67EE88 pKa = 5.5FVWEE92 pKa = 4.34CQQLLRR98 pKa = 11.84DD99 pKa = 3.72GSFDD103 pKa = 2.96IVIYY107 pKa = 10.33YY108 pKa = 9.4RR109 pKa = 11.84ASADD113 pKa = 2.96HH114 pKa = 6.71GAILDD119 pKa = 4.43EE120 pKa = 4.24IQEE123 pKa = 4.27LGFDD127 pKa = 3.62VTGVEE132 pKa = 4.95GEE134 pKa = 4.07
MM1 pKa = 7.85ADD3 pKa = 4.17ALDD6 pKa = 4.18DD7 pKa = 5.97DD8 pKa = 4.6IYY10 pKa = 11.48QRR12 pKa = 11.84TKK14 pKa = 11.12ALLEE18 pKa = 4.39PGEE21 pKa = 4.25IKK23 pKa = 11.07LNGAVVHH30 pKa = 5.86TEE32 pKa = 3.62YY33 pKa = 11.18DD34 pKa = 3.22GGDD37 pKa = 3.85DD38 pKa = 3.74VKK40 pKa = 11.01MMQATIDD47 pKa = 3.66VGDD50 pKa = 4.23VIAAHH55 pKa = 6.52SGHH58 pKa = 7.2DD59 pKa = 3.65PNDD62 pKa = 3.68CYY64 pKa = 11.34VYY66 pKa = 10.73SGNDD70 pKa = 3.42DD71 pKa = 4.02PDD73 pKa = 3.96FSSNQHH79 pKa = 5.89QGLTLDD85 pKa = 4.21DD86 pKa = 4.98EE87 pKa = 4.67EE88 pKa = 5.5FVWEE92 pKa = 4.34CQQLLRR98 pKa = 11.84DD99 pKa = 3.72GSFDD103 pKa = 2.96IVIYY107 pKa = 10.33YY108 pKa = 9.4RR109 pKa = 11.84ASADD113 pKa = 2.96HH114 pKa = 6.71GAILDD119 pKa = 4.43EE120 pKa = 4.24IQEE123 pKa = 4.27LGFDD127 pKa = 3.62VTGVEE132 pKa = 4.95GEE134 pKa = 4.07
Molecular weight: 14.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G4GCB0|G4GCB0_NATGS Cupin OS=Natronobacterium gregoryi (strain ATCC 43098 / DSM 3393 / CCM 3738 / CIP 104747 / IAM 13177 / JCM 8860 / NBRC 102187 / NCIMB 2189 / SP2) OX=797304 GN=Natgr_0271 PE=4 SV=1
MM1 pKa = 7.28EE2 pKa = 4.87RR3 pKa = 11.84WQRR6 pKa = 11.84RR7 pKa = 11.84AVKK10 pKa = 8.78TITGIVVLVFLTSLAYY26 pKa = 9.66HH27 pKa = 5.82YY28 pKa = 11.56VMIVFEE34 pKa = 4.86GRR36 pKa = 11.84SSSYY40 pKa = 9.67FHH42 pKa = 6.25STQVVVEE49 pKa = 4.26TFPGTGYY56 pKa = 10.99GSDD59 pKa = 4.3SPWEE63 pKa = 4.13SAVGNLFVIVMGLSTFCSCLSFSRR87 pKa = 11.84TSSGPSSSGRR97 pKa = 11.84SRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84YY102 pKa = 8.46RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84SSRR109 pKa = 11.84ITSWCVATAPASNDD123 pKa = 3.34SASEE127 pKa = 4.03TKK129 pKa = 10.4ARR131 pKa = 11.84QAGADD136 pKa = 3.82YY137 pKa = 10.99VLSLPDD143 pKa = 2.99ISGRR147 pKa = 11.84LLALDD152 pKa = 3.76VLRR155 pKa = 11.84DD156 pKa = 3.5ASISYY161 pKa = 9.99DD162 pKa = 3.34RR163 pKa = 11.84QLKK166 pKa = 9.36IVRR169 pKa = 11.84FEE171 pKa = 4.41ADD173 pKa = 3.5PLTGRR178 pKa = 11.84AVTNTPLPEE187 pKa = 4.47LNCTLIAVEE196 pKa = 4.73RR197 pKa = 11.84SDD199 pKa = 5.3EE200 pKa = 4.21NVTDD204 pKa = 4.47PAPSPISGAATTCCSPVTTTRR225 pKa = 11.84STRR228 pKa = 11.84SNQNSSKK235 pKa = 10.09FSSVVAATVRR245 pKa = 11.84WRR247 pKa = 11.84KK248 pKa = 8.91YY249 pKa = 8.86RR250 pKa = 11.84RR251 pKa = 11.84CSSSVLTAVFATRR264 pKa = 11.84HH265 pKa = 4.81YY266 pKa = 11.03RR267 pKa = 11.84NDD269 pKa = 3.37LTVHH273 pKa = 6.48LGVTPSEE280 pKa = 4.1NLRR283 pKa = 11.84FAWVLEE289 pKa = 4.31TNDD292 pKa = 2.93FRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84CGFRR300 pKa = 11.84VVGPASITRR309 pKa = 11.84SAVAQLGRR317 pKa = 11.84SSLLSARR324 pKa = 11.84SVRR327 pKa = 11.84LVPSQAA333 pKa = 3.25
MM1 pKa = 7.28EE2 pKa = 4.87RR3 pKa = 11.84WQRR6 pKa = 11.84RR7 pKa = 11.84AVKK10 pKa = 8.78TITGIVVLVFLTSLAYY26 pKa = 9.66HH27 pKa = 5.82YY28 pKa = 11.56VMIVFEE34 pKa = 4.86GRR36 pKa = 11.84SSSYY40 pKa = 9.67FHH42 pKa = 6.25STQVVVEE49 pKa = 4.26TFPGTGYY56 pKa = 10.99GSDD59 pKa = 4.3SPWEE63 pKa = 4.13SAVGNLFVIVMGLSTFCSCLSFSRR87 pKa = 11.84TSSGPSSSGRR97 pKa = 11.84SRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84YY102 pKa = 8.46RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84SSRR109 pKa = 11.84ITSWCVATAPASNDD123 pKa = 3.34SASEE127 pKa = 4.03TKK129 pKa = 10.4ARR131 pKa = 11.84QAGADD136 pKa = 3.82YY137 pKa = 10.99VLSLPDD143 pKa = 2.99ISGRR147 pKa = 11.84LLALDD152 pKa = 3.76VLRR155 pKa = 11.84DD156 pKa = 3.5ASISYY161 pKa = 9.99DD162 pKa = 3.34RR163 pKa = 11.84QLKK166 pKa = 9.36IVRR169 pKa = 11.84FEE171 pKa = 4.41ADD173 pKa = 3.5PLTGRR178 pKa = 11.84AVTNTPLPEE187 pKa = 4.47LNCTLIAVEE196 pKa = 4.73RR197 pKa = 11.84SDD199 pKa = 5.3EE200 pKa = 4.21NVTDD204 pKa = 4.47PAPSPISGAATTCCSPVTTTRR225 pKa = 11.84STRR228 pKa = 11.84SNQNSSKK235 pKa = 10.09FSSVVAATVRR245 pKa = 11.84WRR247 pKa = 11.84KK248 pKa = 8.91YY249 pKa = 8.86RR250 pKa = 11.84RR251 pKa = 11.84CSSSVLTAVFATRR264 pKa = 11.84HH265 pKa = 4.81YY266 pKa = 11.03RR267 pKa = 11.84NDD269 pKa = 3.37LTVHH273 pKa = 6.48LGVTPSEE280 pKa = 4.1NLRR283 pKa = 11.84FAWVLEE289 pKa = 4.31TNDD292 pKa = 2.93FRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84CGFRR300 pKa = 11.84VVGPASITRR309 pKa = 11.84SAVAQLGRR317 pKa = 11.84SSLLSARR324 pKa = 11.84SVRR327 pKa = 11.84LVPSQAA333 pKa = 3.25
Molecular weight: 36.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1028101 |
29 |
1681 |
283.7 |
31.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.17 ± 0.053 | 0.783 ± 0.014 |
8.716 ± 0.056 | 9.587 ± 0.062 |
3.228 ± 0.031 | 8.068 ± 0.042 |
1.987 ± 0.018 | 4.24 ± 0.036 |
1.859 ± 0.024 | 8.932 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.68 ± 0.02 | 2.254 ± 0.019 |
4.539 ± 0.029 | 2.554 ± 0.026 |
6.41 ± 0.042 | 5.552 ± 0.03 |
6.493 ± 0.035 | 9.098 ± 0.041 |
1.113 ± 0.015 | 2.734 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
