
Cellulomonas shaoxiangyii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Cellulomonas
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
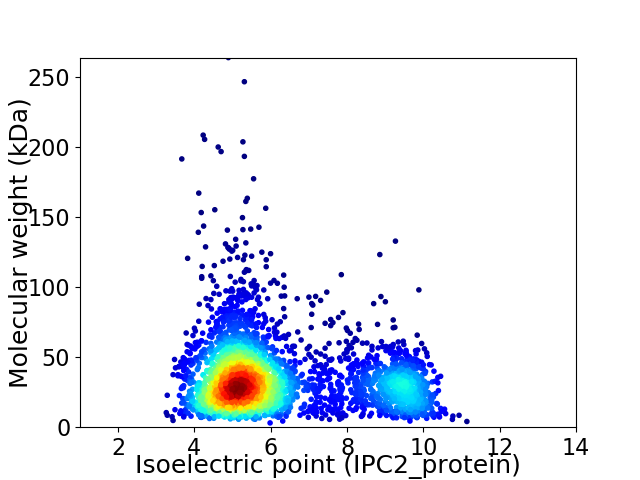
Virtual 2D-PAGE plot for 3337 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1CME5|A0A4V1CME5_9CELL GNAT family N-acetyltransferase OS=Cellulomonas shaoxiangyii OX=2566013 GN=E5225_02975 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 10.29RR3 pKa = 11.84IQRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84GLTLAASVAGLGLVLTACSGGDD30 pKa = 3.44SGDD33 pKa = 3.82GGDD36 pKa = 4.22GGTTGGAGEE45 pKa = 4.24AAVDD49 pKa = 3.49CSEE52 pKa = 3.95FEE54 pKa = 4.15EE55 pKa = 4.76YY56 pKa = 11.18GDD58 pKa = 4.79LEE60 pKa = 4.56GTTVTVYY67 pKa = 10.41TSIVDD72 pKa = 3.77PEE74 pKa = 4.38AEE76 pKa = 4.14QQRR79 pKa = 11.84DD80 pKa = 3.32SYY82 pKa = 12.02APFEE86 pKa = 4.36QCTGATIEE94 pKa = 4.29YY95 pKa = 8.59EE96 pKa = 4.15GSRR99 pKa = 11.84EE100 pKa = 3.93FEE102 pKa = 3.84AQLPVRR108 pKa = 11.84LEE110 pKa = 4.11AGNPPDD116 pKa = 3.65IAYY119 pKa = 9.42IPQPGFLASLVAEE132 pKa = 4.45YY133 pKa = 10.42PDD135 pKa = 3.66VVQAAPEE142 pKa = 4.02AVVDD146 pKa = 4.03NANEE150 pKa = 4.26YY151 pKa = 10.9YY152 pKa = 10.32SEE154 pKa = 3.96EE155 pKa = 3.94WVNYY159 pKa = 7.04GTVDD163 pKa = 3.13GTLYY167 pKa = 9.35ATPLGANVKK176 pKa = 10.15SFVWYY181 pKa = 10.36SPALFEE187 pKa = 4.65EE188 pKa = 4.38NGYY191 pKa = 9.86EE192 pKa = 5.91IPATWQDD199 pKa = 4.2LLDD202 pKa = 4.06LTDD205 pKa = 5.55QIVEE209 pKa = 4.61DD210 pKa = 4.48NPDD213 pKa = 3.55AKK215 pKa = 9.93PWCAGIEE222 pKa = 4.21SGDD225 pKa = 3.58ATGWPATDD233 pKa = 3.11WLEE236 pKa = 4.41DD237 pKa = 3.59VVLRR241 pKa = 11.84TAGPDD246 pKa = 4.24VYY248 pKa = 10.79DD249 pKa = 2.82QWVNHH254 pKa = 7.0EE255 pKa = 4.78IPFDD259 pKa = 4.16DD260 pKa = 4.3PQIVAALEE268 pKa = 3.92EE269 pKa = 4.33AGGILKK275 pKa = 10.62NEE277 pKa = 4.49DD278 pKa = 3.73YY279 pKa = 11.83VNGGLGNVEE288 pKa = 5.09SIATAPWTEE297 pKa = 3.78AGLPVIDD304 pKa = 5.06GSCFLHH310 pKa = 6.93RR311 pKa = 11.84AANFYY316 pKa = 10.49AANLEE321 pKa = 4.3GYY323 pKa = 9.09EE324 pKa = 4.05VAEE327 pKa = 4.95DD328 pKa = 4.1GEE330 pKa = 4.45IFAFYY335 pKa = 10.71LPGPTEE341 pKa = 4.08DD342 pKa = 4.21EE343 pKa = 4.35KK344 pKa = 11.35PLLGGGEE351 pKa = 4.35FVTAFSDD358 pKa = 3.76RR359 pKa = 11.84PEE361 pKa = 3.99VQAFHH366 pKa = 7.45AYY368 pKa = 10.19LSSPQWANAKK378 pKa = 10.37AEE380 pKa = 4.23STPGGGWLSANSGLDD395 pKa = 3.26RR396 pKa = 11.84DD397 pKa = 4.84LLTSPIDD404 pKa = 3.45QLSFDD409 pKa = 5.11LLTDD413 pKa = 3.55DD414 pKa = 5.54AYY416 pKa = 9.96TFRR419 pKa = 11.84FDD421 pKa = 5.99GSDD424 pKa = 3.49QMPGAVGAGAFWRR437 pKa = 11.84EE438 pKa = 3.62MTAWISPGQDD448 pKa = 2.75AQTTLTNIEE457 pKa = 4.31AAWPTSS463 pKa = 3.27
MM1 pKa = 7.46KK2 pKa = 10.29RR3 pKa = 11.84IQRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84GLTLAASVAGLGLVLTACSGGDD30 pKa = 3.44SGDD33 pKa = 3.82GGDD36 pKa = 4.22GGTTGGAGEE45 pKa = 4.24AAVDD49 pKa = 3.49CSEE52 pKa = 3.95FEE54 pKa = 4.15EE55 pKa = 4.76YY56 pKa = 11.18GDD58 pKa = 4.79LEE60 pKa = 4.56GTTVTVYY67 pKa = 10.41TSIVDD72 pKa = 3.77PEE74 pKa = 4.38AEE76 pKa = 4.14QQRR79 pKa = 11.84DD80 pKa = 3.32SYY82 pKa = 12.02APFEE86 pKa = 4.36QCTGATIEE94 pKa = 4.29YY95 pKa = 8.59EE96 pKa = 4.15GSRR99 pKa = 11.84EE100 pKa = 3.93FEE102 pKa = 3.84AQLPVRR108 pKa = 11.84LEE110 pKa = 4.11AGNPPDD116 pKa = 3.65IAYY119 pKa = 9.42IPQPGFLASLVAEE132 pKa = 4.45YY133 pKa = 10.42PDD135 pKa = 3.66VVQAAPEE142 pKa = 4.02AVVDD146 pKa = 4.03NANEE150 pKa = 4.26YY151 pKa = 10.9YY152 pKa = 10.32SEE154 pKa = 3.96EE155 pKa = 3.94WVNYY159 pKa = 7.04GTVDD163 pKa = 3.13GTLYY167 pKa = 9.35ATPLGANVKK176 pKa = 10.15SFVWYY181 pKa = 10.36SPALFEE187 pKa = 4.65EE188 pKa = 4.38NGYY191 pKa = 9.86EE192 pKa = 5.91IPATWQDD199 pKa = 4.2LLDD202 pKa = 4.06LTDD205 pKa = 5.55QIVEE209 pKa = 4.61DD210 pKa = 4.48NPDD213 pKa = 3.55AKK215 pKa = 9.93PWCAGIEE222 pKa = 4.21SGDD225 pKa = 3.58ATGWPATDD233 pKa = 3.11WLEE236 pKa = 4.41DD237 pKa = 3.59VVLRR241 pKa = 11.84TAGPDD246 pKa = 4.24VYY248 pKa = 10.79DD249 pKa = 2.82QWVNHH254 pKa = 7.0EE255 pKa = 4.78IPFDD259 pKa = 4.16DD260 pKa = 4.3PQIVAALEE268 pKa = 3.92EE269 pKa = 4.33AGGILKK275 pKa = 10.62NEE277 pKa = 4.49DD278 pKa = 3.73YY279 pKa = 11.83VNGGLGNVEE288 pKa = 5.09SIATAPWTEE297 pKa = 3.78AGLPVIDD304 pKa = 5.06GSCFLHH310 pKa = 6.93RR311 pKa = 11.84AANFYY316 pKa = 10.49AANLEE321 pKa = 4.3GYY323 pKa = 9.09EE324 pKa = 4.05VAEE327 pKa = 4.95DD328 pKa = 4.1GEE330 pKa = 4.45IFAFYY335 pKa = 10.71LPGPTEE341 pKa = 4.08DD342 pKa = 4.21EE343 pKa = 4.35KK344 pKa = 11.35PLLGGGEE351 pKa = 4.35FVTAFSDD358 pKa = 3.76RR359 pKa = 11.84PEE361 pKa = 3.99VQAFHH366 pKa = 7.45AYY368 pKa = 10.19LSSPQWANAKK378 pKa = 10.37AEE380 pKa = 4.23STPGGGWLSANSGLDD395 pKa = 3.26RR396 pKa = 11.84DD397 pKa = 4.84LLTSPIDD404 pKa = 3.45QLSFDD409 pKa = 5.11LLTDD413 pKa = 3.55DD414 pKa = 5.54AYY416 pKa = 9.96TFRR419 pKa = 11.84FDD421 pKa = 5.99GSDD424 pKa = 3.49QMPGAVGAGAFWRR437 pKa = 11.84EE438 pKa = 3.62MTAWISPGQDD448 pKa = 2.75AQTTLTNIEE457 pKa = 4.31AAWPTSS463 pKa = 3.27
Molecular weight: 49.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P7SLV9|A0A4P7SLV9_9CELL ArsR family transcriptional regulator OS=Cellulomonas shaoxiangyii OX=2566013 GN=E5225_10215 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1147658 |
29 |
2544 |
343.9 |
36.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.224 ± 0.068 | 0.553 ± 0.01 |
6.6 ± 0.033 | 4.992 ± 0.042 |
2.341 ± 0.026 | 9.67 ± 0.039 |
2.163 ± 0.019 | 2.303 ± 0.033 |
1.15 ± 0.024 | 10.244 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.445 ± 0.016 | 1.375 ± 0.021 |
6.318 ± 0.036 | 2.534 ± 0.022 |
8.279 ± 0.043 | 4.407 ± 0.026 |
6.391 ± 0.042 | 10.632 ± 0.045 |
1.611 ± 0.019 | 1.769 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
