
Aleutian mink disease parvovirus (strain G) (ADV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Parvovirinae; Amdoparvovirus; Carnivore amdoparvovirus 1; Aleutian mink disease virus
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
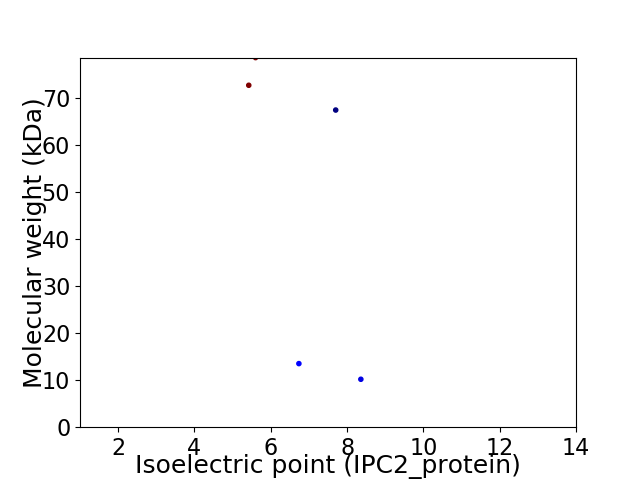
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
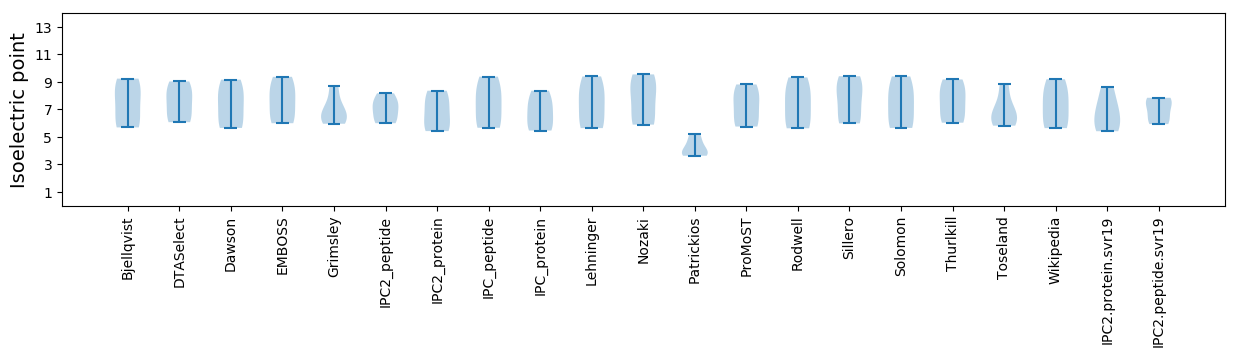
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P24029-2|CAPSD-2_ADVG Isoform of P24029 Isoform VP2 of Capsid protein VP1 OS=Aleutian mink disease parvovirus (strain G) OX=10783 PE=4 SV=3
MM1 pKa = 7.61DD2 pKa = 4.39TEE4 pKa = 4.06QATNQTAEE12 pKa = 4.04AGGGGGGGGGGGGGGGGVGNSTGGFNNTTEE42 pKa = 4.13FKK44 pKa = 10.82VINNEE49 pKa = 4.17VYY51 pKa = 8.79ITCHH55 pKa = 5.26ATRR58 pKa = 11.84MVHH61 pKa = 6.78INQADD66 pKa = 3.58TDD68 pKa = 3.77EE69 pKa = 4.15YY70 pKa = 11.37LIFNAGRR77 pKa = 11.84TTDD80 pKa = 3.58TKK82 pKa = 10.25THH84 pKa = 4.52QQKK87 pKa = 10.94LNLEE91 pKa = 4.21FFVYY95 pKa = 10.6DD96 pKa = 4.2DD97 pKa = 3.63FHH99 pKa = 7.41QQVMTPWYY107 pKa = 9.87IVDD110 pKa = 3.74SNAWGVWMSPKK121 pKa = 10.4DD122 pKa = 3.74FQQMKK127 pKa = 8.04TLCSEE132 pKa = 4.05ISLVTLEE139 pKa = 4.18QEE141 pKa = 3.92IDD143 pKa = 3.25NVTIKK148 pKa = 9.97TVTEE152 pKa = 4.31TNQGNASTKK161 pKa = 10.09QFNNDD166 pKa = 2.95LTASLQVALDD176 pKa = 3.76TNNILPYY183 pKa = 10.23TPAAPLGEE191 pKa = 4.19TLGFVPWRR199 pKa = 11.84ATKK202 pKa = 8.31PTQYY206 pKa = 10.37RR207 pKa = 11.84YY208 pKa = 9.54YY209 pKa = 9.47HH210 pKa = 6.5PCYY213 pKa = 10.07IYY215 pKa = 10.9NRR217 pKa = 11.84YY218 pKa = 9.47PNIQKK223 pKa = 10.0VATEE227 pKa = 4.13TLTWDD232 pKa = 3.81AVQDD236 pKa = 4.13DD237 pKa = 4.76YY238 pKa = 12.2LSVDD242 pKa = 3.22EE243 pKa = 5.11QYY245 pKa = 11.6FNFITIEE252 pKa = 3.84NNIPINILRR261 pKa = 11.84TGDD264 pKa = 3.51NFHH267 pKa = 6.66TGLYY271 pKa = 9.22EE272 pKa = 4.78FNSKK276 pKa = 9.35PCKK279 pKa = 9.38LTLSYY284 pKa = 11.09QSTRR288 pKa = 11.84CLGLPPLCKK297 pKa = 9.94PKK299 pKa = 10.35TDD301 pKa = 3.56TTHH304 pKa = 7.33KK305 pKa = 9.74VTSKK309 pKa = 11.17EE310 pKa = 3.79NGADD314 pKa = 4.09LIYY317 pKa = 10.59IQGQDD322 pKa = 3.31NTRR325 pKa = 11.84LGHH328 pKa = 5.84FWGEE332 pKa = 3.7EE333 pKa = 3.69RR334 pKa = 11.84GKK336 pKa = 11.06KK337 pKa = 7.95NAEE340 pKa = 3.93MNRR343 pKa = 11.84IRR345 pKa = 11.84PYY347 pKa = 10.72NIGYY351 pKa = 9.48QYY353 pKa = 10.45PEE355 pKa = 4.2WIIPAGLQGSYY366 pKa = 10.22FAGGPRR372 pKa = 11.84QWSDD376 pKa = 3.02TTKK379 pKa = 10.85GAGTHH384 pKa = 5.4SQHH387 pKa = 5.97LQQNFSTRR395 pKa = 11.84YY396 pKa = 9.3IYY398 pKa = 10.51DD399 pKa = 3.62RR400 pKa = 11.84NHH402 pKa = 6.48GGDD405 pKa = 3.75NEE407 pKa = 4.03VDD409 pKa = 3.96LLDD412 pKa = 5.26GIPIHH417 pKa = 6.38EE418 pKa = 4.43RR419 pKa = 11.84SNYY422 pKa = 9.77YY423 pKa = 10.19SDD425 pKa = 5.18NEE427 pKa = 4.33IEE429 pKa = 4.14QHH431 pKa = 5.1TAKK434 pKa = 10.19QPKK437 pKa = 9.71LRR439 pKa = 11.84TPPIHH444 pKa = 6.73HH445 pKa = 6.74SKK447 pKa = 9.66IDD449 pKa = 3.27SWEE452 pKa = 4.09EE453 pKa = 3.79EE454 pKa = 4.21GWPAASGTHH463 pKa = 6.32FEE465 pKa = 5.02DD466 pKa = 3.61EE467 pKa = 4.8VIYY470 pKa = 10.85LDD472 pKa = 3.58YY473 pKa = 11.23FNFSGEE479 pKa = 4.03QEE481 pKa = 4.33LNFPHH486 pKa = 6.93EE487 pKa = 4.47VLDD490 pKa = 5.59DD491 pKa = 3.87AAQMKK496 pKa = 10.23KK497 pKa = 10.43LLNSYY502 pKa = 10.11QPTVAQDD509 pKa = 3.57NVGPVYY515 pKa = 10.22PWGQIWDD522 pKa = 4.16KK523 pKa = 10.56KK524 pKa = 9.64PHH526 pKa = 6.31MDD528 pKa = 4.78HH529 pKa = 6.78KK530 pKa = 11.06PSMNNNAPFVCKK542 pKa = 10.44NNPPGQLFVKK552 pKa = 9.57LTEE555 pKa = 4.26NLTDD559 pKa = 3.34TFNYY563 pKa = 10.31DD564 pKa = 3.53EE565 pKa = 4.8NPDD568 pKa = 3.81RR569 pKa = 11.84IKK571 pKa = 10.22TYY573 pKa = 10.59GYY575 pKa = 7.33FTWRR579 pKa = 11.84GKK581 pKa = 10.53LVLKK585 pKa = 10.79GKK587 pKa = 10.28LSQVTCWNPVKK598 pKa = 10.52RR599 pKa = 11.84EE600 pKa = 4.15LIGEE604 pKa = 4.24PGVFTKK610 pKa = 10.59DD611 pKa = 3.42KK612 pKa = 9.94YY613 pKa = 10.84HH614 pKa = 6.71KK615 pKa = 10.18QIPNNKK621 pKa = 8.58GNFEE625 pKa = 4.05IGLQYY630 pKa = 10.86GRR632 pKa = 11.84STIKK636 pKa = 10.69YY637 pKa = 8.68IYY639 pKa = 10.37
MM1 pKa = 7.61DD2 pKa = 4.39TEE4 pKa = 4.06QATNQTAEE12 pKa = 4.04AGGGGGGGGGGGGGGGGVGNSTGGFNNTTEE42 pKa = 4.13FKK44 pKa = 10.82VINNEE49 pKa = 4.17VYY51 pKa = 8.79ITCHH55 pKa = 5.26ATRR58 pKa = 11.84MVHH61 pKa = 6.78INQADD66 pKa = 3.58TDD68 pKa = 3.77EE69 pKa = 4.15YY70 pKa = 11.37LIFNAGRR77 pKa = 11.84TTDD80 pKa = 3.58TKK82 pKa = 10.25THH84 pKa = 4.52QQKK87 pKa = 10.94LNLEE91 pKa = 4.21FFVYY95 pKa = 10.6DD96 pKa = 4.2DD97 pKa = 3.63FHH99 pKa = 7.41QQVMTPWYY107 pKa = 9.87IVDD110 pKa = 3.74SNAWGVWMSPKK121 pKa = 10.4DD122 pKa = 3.74FQQMKK127 pKa = 8.04TLCSEE132 pKa = 4.05ISLVTLEE139 pKa = 4.18QEE141 pKa = 3.92IDD143 pKa = 3.25NVTIKK148 pKa = 9.97TVTEE152 pKa = 4.31TNQGNASTKK161 pKa = 10.09QFNNDD166 pKa = 2.95LTASLQVALDD176 pKa = 3.76TNNILPYY183 pKa = 10.23TPAAPLGEE191 pKa = 4.19TLGFVPWRR199 pKa = 11.84ATKK202 pKa = 8.31PTQYY206 pKa = 10.37RR207 pKa = 11.84YY208 pKa = 9.54YY209 pKa = 9.47HH210 pKa = 6.5PCYY213 pKa = 10.07IYY215 pKa = 10.9NRR217 pKa = 11.84YY218 pKa = 9.47PNIQKK223 pKa = 10.0VATEE227 pKa = 4.13TLTWDD232 pKa = 3.81AVQDD236 pKa = 4.13DD237 pKa = 4.76YY238 pKa = 12.2LSVDD242 pKa = 3.22EE243 pKa = 5.11QYY245 pKa = 11.6FNFITIEE252 pKa = 3.84NNIPINILRR261 pKa = 11.84TGDD264 pKa = 3.51NFHH267 pKa = 6.66TGLYY271 pKa = 9.22EE272 pKa = 4.78FNSKK276 pKa = 9.35PCKK279 pKa = 9.38LTLSYY284 pKa = 11.09QSTRR288 pKa = 11.84CLGLPPLCKK297 pKa = 9.94PKK299 pKa = 10.35TDD301 pKa = 3.56TTHH304 pKa = 7.33KK305 pKa = 9.74VTSKK309 pKa = 11.17EE310 pKa = 3.79NGADD314 pKa = 4.09LIYY317 pKa = 10.59IQGQDD322 pKa = 3.31NTRR325 pKa = 11.84LGHH328 pKa = 5.84FWGEE332 pKa = 3.7EE333 pKa = 3.69RR334 pKa = 11.84GKK336 pKa = 11.06KK337 pKa = 7.95NAEE340 pKa = 3.93MNRR343 pKa = 11.84IRR345 pKa = 11.84PYY347 pKa = 10.72NIGYY351 pKa = 9.48QYY353 pKa = 10.45PEE355 pKa = 4.2WIIPAGLQGSYY366 pKa = 10.22FAGGPRR372 pKa = 11.84QWSDD376 pKa = 3.02TTKK379 pKa = 10.85GAGTHH384 pKa = 5.4SQHH387 pKa = 5.97LQQNFSTRR395 pKa = 11.84YY396 pKa = 9.3IYY398 pKa = 10.51DD399 pKa = 3.62RR400 pKa = 11.84NHH402 pKa = 6.48GGDD405 pKa = 3.75NEE407 pKa = 4.03VDD409 pKa = 3.96LLDD412 pKa = 5.26GIPIHH417 pKa = 6.38EE418 pKa = 4.43RR419 pKa = 11.84SNYY422 pKa = 9.77YY423 pKa = 10.19SDD425 pKa = 5.18NEE427 pKa = 4.33IEE429 pKa = 4.14QHH431 pKa = 5.1TAKK434 pKa = 10.19QPKK437 pKa = 9.71LRR439 pKa = 11.84TPPIHH444 pKa = 6.73HH445 pKa = 6.74SKK447 pKa = 9.66IDD449 pKa = 3.27SWEE452 pKa = 4.09EE453 pKa = 3.79EE454 pKa = 4.21GWPAASGTHH463 pKa = 6.32FEE465 pKa = 5.02DD466 pKa = 3.61EE467 pKa = 4.8VIYY470 pKa = 10.85LDD472 pKa = 3.58YY473 pKa = 11.23FNFSGEE479 pKa = 4.03QEE481 pKa = 4.33LNFPHH486 pKa = 6.93EE487 pKa = 4.47VLDD490 pKa = 5.59DD491 pKa = 3.87AAQMKK496 pKa = 10.23KK497 pKa = 10.43LLNSYY502 pKa = 10.11QPTVAQDD509 pKa = 3.57NVGPVYY515 pKa = 10.22PWGQIWDD522 pKa = 4.16KK523 pKa = 10.56KK524 pKa = 9.64PHH526 pKa = 6.31MDD528 pKa = 4.78HH529 pKa = 6.78KK530 pKa = 11.06PSMNNNAPFVCKK542 pKa = 10.44NNPPGQLFVKK552 pKa = 9.57LTEE555 pKa = 4.26NLTDD559 pKa = 3.34TFNYY563 pKa = 10.31DD564 pKa = 3.53EE565 pKa = 4.8NPDD568 pKa = 3.81RR569 pKa = 11.84IKK571 pKa = 10.22TYY573 pKa = 10.59GYY575 pKa = 7.33FTWRR579 pKa = 11.84GKK581 pKa = 10.53LVLKK585 pKa = 10.79GKK587 pKa = 10.28LSQVTCWNPVKK598 pKa = 10.52RR599 pKa = 11.84EE600 pKa = 4.15LIGEE604 pKa = 4.24PGVFTKK610 pKa = 10.59DD611 pKa = 3.42KK612 pKa = 9.94YY613 pKa = 10.84HH614 pKa = 6.71KK615 pKa = 10.18QIPNNKK621 pKa = 8.58GNFEE625 pKa = 4.05IGLQYY630 pKa = 10.86GRR632 pKa = 11.84STIKK636 pKa = 10.69YY637 pKa = 8.68IYY639 pKa = 10.37
Molecular weight: 72.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P24029-2|CAPSD-2_ADVG Isoform of P24029 Isoform VP2 of Capsid protein VP1 OS=Aleutian mink disease parvovirus (strain G) OX=10783 PE=4 SV=3
MM1 pKa = 7.37AQAQIDD7 pKa = 4.1EE8 pKa = 4.16QRR10 pKa = 11.84RR11 pKa = 11.84LQDD14 pKa = 4.46LYY16 pKa = 11.42VQLKK20 pKa = 10.4KK21 pKa = 10.66EE22 pKa = 4.2INDD25 pKa = 4.01GEE27 pKa = 4.53GVAWLFQQKK36 pKa = 8.97TYY38 pKa = 9.6TDD40 pKa = 4.0KK41 pKa = 11.47DD42 pKa = 3.7NKK44 pKa = 7.85PTKK47 pKa = 9.25ATPPLRR53 pKa = 11.84TTSSDD58 pKa = 5.26LRR60 pKa = 11.84LCDD63 pKa = 4.07CNKK66 pKa = 9.53QHH68 pKa = 6.49QHH70 pKa = 5.05NQSNCWMCGNKK81 pKa = 9.57RR82 pKa = 11.84SRR84 pKa = 11.84RR85 pKa = 11.84ATT87 pKa = 3.29
MM1 pKa = 7.37AQAQIDD7 pKa = 4.1EE8 pKa = 4.16QRR10 pKa = 11.84RR11 pKa = 11.84LQDD14 pKa = 4.46LYY16 pKa = 11.42VQLKK20 pKa = 10.4KK21 pKa = 10.66EE22 pKa = 4.2INDD25 pKa = 4.01GEE27 pKa = 4.53GVAWLFQQKK36 pKa = 8.97TYY38 pKa = 9.6TDD40 pKa = 4.0KK41 pKa = 11.47DD42 pKa = 3.7NKK44 pKa = 7.85PTKK47 pKa = 9.25ATPPLRR53 pKa = 11.84TTSSDD58 pKa = 5.26LRR60 pKa = 11.84LCDD63 pKa = 4.07CNKK66 pKa = 9.53QHH68 pKa = 6.49QHH70 pKa = 5.05NQSNCWMCGNKK81 pKa = 9.57RR82 pKa = 11.84SRR84 pKa = 11.84RR85 pKa = 11.84ATT87 pKa = 3.29
Molecular weight: 10.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2120 |
87 |
690 |
424.0 |
48.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.528 ± 0.204 | 2.075 ± 0.52 |
6.321 ± 0.408 | 5.755 ± 0.364 |
3.443 ± 0.316 | 7.264 ± 0.763 |
2.83 ± 0.321 | 5.566 ± 0.428 |
7.972 ± 1.14 | 6.934 ± 0.509 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.745 ± 0.216 | 7.217 ± 0.378 |
5.0 ± 0.426 | 6.085 ± 0.447 |
3.396 ± 0.328 | 4.151 ± 0.127 |
8.679 ± 0.087 | 4.245 ± 0.223 |
2.217 ± 0.078 | 4.575 ± 0.594 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
