
Avian metapneumovirus (isolate Canada goose/Minnesota/15a/2001) (AMPV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Pneumoviridae; Metapneumovirus; Avian metapneumovirus
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
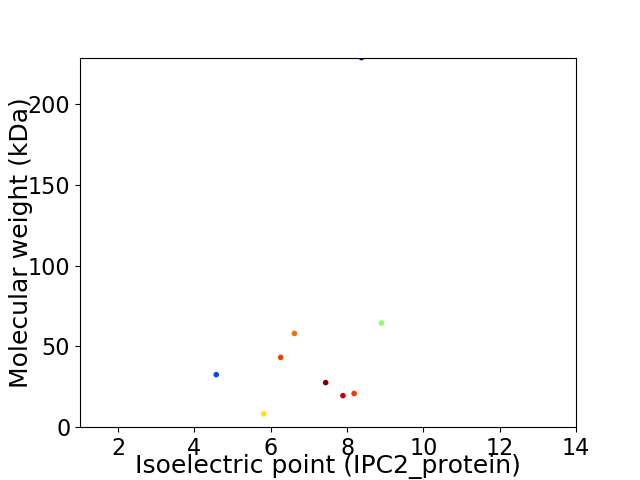
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>sp|Q2Y2M6|NCAP_AMPV1 Nucleoprotein OS=Avian metapneumovirus (isolate Canada goose/Minnesota/15a/2001) OX=652954 GN=N PE=3 SV=1
MM1 pKa = 7.55SFPEE5 pKa = 4.62GKK7 pKa = 10.16DD8 pKa = 2.89ILLMGNEE15 pKa = 3.87AAKK18 pKa = 10.35AAEE21 pKa = 4.13AFQRR25 pKa = 11.84SLKK28 pKa = 10.57KK29 pKa = 9.59IGHH32 pKa = 6.76RR33 pKa = 11.84RR34 pKa = 11.84TQSIVGDD41 pKa = 3.83KK42 pKa = 10.32IITVSEE48 pKa = 4.34TVEE51 pKa = 4.09KK52 pKa = 9.04PTISKK57 pKa = 8.09STKK60 pKa = 8.33VTTPPEE66 pKa = 3.77RR67 pKa = 11.84RR68 pKa = 11.84NAWGEE73 pKa = 4.06KK74 pKa = 9.81PDD76 pKa = 3.46TTRR79 pKa = 11.84NQTEE83 pKa = 3.99EE84 pKa = 4.05ARR86 pKa = 11.84NEE88 pKa = 3.78ATLEE92 pKa = 4.11DD93 pKa = 3.77TSRR96 pKa = 11.84LYY98 pKa = 11.16EE99 pKa = 4.07EE100 pKa = 5.18VFAPTSDD107 pKa = 2.77GKK109 pKa = 9.05TPAEE113 pKa = 4.2EE114 pKa = 4.68GMEE117 pKa = 4.18TPEE120 pKa = 4.11KK121 pKa = 9.79PKK123 pKa = 10.94KK124 pKa = 10.23KK125 pKa = 9.2VTFKK129 pKa = 10.6NDD131 pKa = 2.56EE132 pKa = 4.08SGRR135 pKa = 11.84YY136 pKa = 6.94TKK138 pKa = 11.07LEE140 pKa = 3.96MEE142 pKa = 4.41ALEE145 pKa = 4.47LLSDD149 pKa = 4.56NEE151 pKa = 4.45DD152 pKa = 3.76DD153 pKa = 4.41DD154 pKa = 6.33AEE156 pKa = 4.67SSVLTFEE163 pKa = 5.05EE164 pKa = 4.99KK165 pKa = 9.88DD166 pKa = 3.38TSALSLEE173 pKa = 4.39ARR175 pKa = 11.84LEE177 pKa = 4.44SIDD180 pKa = 4.38EE181 pKa = 4.26KK182 pKa = 11.33LSMILGLLRR191 pKa = 11.84TLNVATAGPTAARR204 pKa = 11.84DD205 pKa = 4.19GIRR208 pKa = 11.84DD209 pKa = 3.41AMVGLRR215 pKa = 11.84EE216 pKa = 4.09EE217 pKa = 5.15LIADD221 pKa = 4.31IIKK224 pKa = 9.71EE225 pKa = 4.32AKK227 pKa = 9.86GKK229 pKa = 9.31AAEE232 pKa = 4.11MMKK235 pKa = 10.69EE236 pKa = 3.78EE237 pKa = 4.8AKK239 pKa = 10.48QKK241 pKa = 10.9SKK243 pKa = 10.6IGNGSVGLTEE253 pKa = 4.85KK254 pKa = 10.84AKK256 pKa = 9.9EE257 pKa = 3.87LNKK260 pKa = 10.09IVEE263 pKa = 4.67DD264 pKa = 4.06EE265 pKa = 4.41STSGEE270 pKa = 4.17SEE272 pKa = 4.1EE273 pKa = 4.4EE274 pKa = 3.98EE275 pKa = 4.11EE276 pKa = 5.02EE277 pKa = 4.08EE278 pKa = 5.79DD279 pKa = 4.42EE280 pKa = 4.99EE281 pKa = 5.09EE282 pKa = 4.98SNPDD286 pKa = 3.34DD287 pKa = 4.88DD288 pKa = 6.29LYY290 pKa = 11.69SLTMM294 pKa = 4.41
MM1 pKa = 7.55SFPEE5 pKa = 4.62GKK7 pKa = 10.16DD8 pKa = 2.89ILLMGNEE15 pKa = 3.87AAKK18 pKa = 10.35AAEE21 pKa = 4.13AFQRR25 pKa = 11.84SLKK28 pKa = 10.57KK29 pKa = 9.59IGHH32 pKa = 6.76RR33 pKa = 11.84RR34 pKa = 11.84TQSIVGDD41 pKa = 3.83KK42 pKa = 10.32IITVSEE48 pKa = 4.34TVEE51 pKa = 4.09KK52 pKa = 9.04PTISKK57 pKa = 8.09STKK60 pKa = 8.33VTTPPEE66 pKa = 3.77RR67 pKa = 11.84RR68 pKa = 11.84NAWGEE73 pKa = 4.06KK74 pKa = 9.81PDD76 pKa = 3.46TTRR79 pKa = 11.84NQTEE83 pKa = 3.99EE84 pKa = 4.05ARR86 pKa = 11.84NEE88 pKa = 3.78ATLEE92 pKa = 4.11DD93 pKa = 3.77TSRR96 pKa = 11.84LYY98 pKa = 11.16EE99 pKa = 4.07EE100 pKa = 5.18VFAPTSDD107 pKa = 2.77GKK109 pKa = 9.05TPAEE113 pKa = 4.2EE114 pKa = 4.68GMEE117 pKa = 4.18TPEE120 pKa = 4.11KK121 pKa = 9.79PKK123 pKa = 10.94KK124 pKa = 10.23KK125 pKa = 9.2VTFKK129 pKa = 10.6NDD131 pKa = 2.56EE132 pKa = 4.08SGRR135 pKa = 11.84YY136 pKa = 6.94TKK138 pKa = 11.07LEE140 pKa = 3.96MEE142 pKa = 4.41ALEE145 pKa = 4.47LLSDD149 pKa = 4.56NEE151 pKa = 4.45DD152 pKa = 3.76DD153 pKa = 4.41DD154 pKa = 6.33AEE156 pKa = 4.67SSVLTFEE163 pKa = 5.05EE164 pKa = 4.99KK165 pKa = 9.88DD166 pKa = 3.38TSALSLEE173 pKa = 4.39ARR175 pKa = 11.84LEE177 pKa = 4.44SIDD180 pKa = 4.38EE181 pKa = 4.26KK182 pKa = 11.33LSMILGLLRR191 pKa = 11.84TLNVATAGPTAARR204 pKa = 11.84DD205 pKa = 4.19GIRR208 pKa = 11.84DD209 pKa = 3.41AMVGLRR215 pKa = 11.84EE216 pKa = 4.09EE217 pKa = 5.15LIADD221 pKa = 4.31IIKK224 pKa = 9.71EE225 pKa = 4.32AKK227 pKa = 9.86GKK229 pKa = 9.31AAEE232 pKa = 4.11MMKK235 pKa = 10.69EE236 pKa = 3.78EE237 pKa = 4.8AKK239 pKa = 10.48QKK241 pKa = 10.9SKK243 pKa = 10.6IGNGSVGLTEE253 pKa = 4.85KK254 pKa = 10.84AKK256 pKa = 9.9EE257 pKa = 3.87LNKK260 pKa = 10.09IVEE263 pKa = 4.67DD264 pKa = 4.06EE265 pKa = 4.41STSGEE270 pKa = 4.17SEE272 pKa = 4.1EE273 pKa = 4.4EE274 pKa = 3.98EE275 pKa = 4.11EE276 pKa = 5.02EE277 pKa = 4.08EE278 pKa = 5.79DD279 pKa = 4.42EE280 pKa = 4.99EE281 pKa = 5.09EE282 pKa = 4.98SNPDD286 pKa = 3.34DD287 pKa = 4.88DD288 pKa = 6.29LYY290 pKa = 11.69SLTMM294 pKa = 4.41
Molecular weight: 32.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q2Y2M0|SH_AMPV1 Small hydrophobic protein OS=Avian metapneumovirus (isolate Canada goose/Minnesota/15a/2001) OX=652954 GN=SH PE=3 SV=1
MM1 pKa = 7.11EE2 pKa = 4.47VKK4 pKa = 10.42VEE6 pKa = 4.08NVGKK10 pKa = 9.88SQEE13 pKa = 4.18LKK15 pKa = 10.47VKK17 pKa = 9.99VKK19 pKa = 10.96NFIKK23 pKa = 10.62RR24 pKa = 11.84SDD26 pKa = 3.73CKK28 pKa = 10.75KK29 pKa = 10.78KK30 pKa = 10.74LFALILGLVSFEE42 pKa = 3.78LTMNIMLSVMYY53 pKa = 10.62VEE55 pKa = 5.09SNEE58 pKa = 3.96ALSLCRR64 pKa = 11.84IQGTPAPRR72 pKa = 11.84DD73 pKa = 3.29NKK75 pKa = 10.64TNTEE79 pKa = 4.0NTKK82 pKa = 10.62KK83 pKa = 9.2EE84 pKa = 4.5TTFHH88 pKa = 5.8TTTTTRR94 pKa = 11.84DD95 pKa = 3.34PEE97 pKa = 4.07VRR99 pKa = 11.84EE100 pKa = 4.23TKK102 pKa = 6.87TTKK105 pKa = 10.36PKK107 pKa = 9.29TNEE110 pKa = 3.85GATSPSRR117 pKa = 11.84NLTTKK122 pKa = 10.91GDD124 pKa = 3.14IHH126 pKa = 5.48QTTRR130 pKa = 11.84ATTEE134 pKa = 3.89AEE136 pKa = 4.05LEE138 pKa = 4.3KK139 pKa = 10.64QSKK142 pKa = 7.06QTIEE146 pKa = 4.46PDD148 pKa = 3.16TSTKK152 pKa = 10.32KK153 pKa = 8.82HH154 pKa = 5.28TPTRR158 pKa = 11.84PSSEE162 pKa = 4.3SPTTTQATAQLTTPTAPKK180 pKa = 10.6ASIAPKK186 pKa = 10.05NRR188 pKa = 11.84QATTKK193 pKa = 9.16KK194 pKa = 8.79TEE196 pKa = 4.02TGTTTTSRR204 pKa = 11.84AKK206 pKa = 9.28KK207 pKa = 8.3TNNPTEE213 pKa = 4.23TATTTLKK220 pKa = 10.3ATTEE224 pKa = 4.02TGKK227 pKa = 10.92GKK229 pKa = 10.31EE230 pKa = 4.45GPTQHH235 pKa = 6.76TIKK238 pKa = 10.2EE239 pKa = 4.32QPEE242 pKa = 4.17TTAGEE247 pKa = 4.39TTTPQSRR254 pKa = 11.84RR255 pKa = 11.84TTSRR259 pKa = 11.84PAPTTKK265 pKa = 10.09TEE267 pKa = 4.14EE268 pKa = 4.02EE269 pKa = 4.35AEE271 pKa = 4.35TTKK274 pKa = 10.42TRR276 pKa = 11.84TTKK279 pKa = 9.28STQTSTGPPGPTRR292 pKa = 11.84STPSKK297 pKa = 9.71TATEE301 pKa = 4.07NNKK304 pKa = 8.77RR305 pKa = 11.84TTTIKK310 pKa = 10.59RR311 pKa = 11.84PNTANTDD318 pKa = 2.94SRR320 pKa = 11.84QQTRR324 pKa = 11.84TTAEE328 pKa = 3.77QDD330 pKa = 3.16RR331 pKa = 11.84QIQTKK336 pKa = 10.31AKK338 pKa = 7.08PTTNGAHH345 pKa = 6.33AQTTTTPEE353 pKa = 4.22HH354 pKa = 5.87NTDD357 pKa = 3.23TTNSTKK363 pKa = 10.44EE364 pKa = 3.74SSKK367 pKa = 10.64EE368 pKa = 3.86DD369 pKa = 3.16KK370 pKa = 8.18TTRR373 pKa = 11.84DD374 pKa = 3.8PSSKK378 pKa = 9.72TPTDD382 pKa = 3.26QEE384 pKa = 4.34DD385 pKa = 3.6ASKK388 pKa = 9.6GTTAANPRR396 pKa = 11.84KK397 pKa = 8.46NTEE400 pKa = 4.0ANTRR404 pKa = 11.84TPPTTTPTRR413 pKa = 11.84HH414 pKa = 4.38TTEE417 pKa = 4.32SATSTTGDD425 pKa = 3.04KK426 pKa = 10.44TKK428 pKa = 11.14AKK430 pKa = 4.72TTRR433 pKa = 11.84WKK435 pKa = 9.48STADD439 pKa = 3.48RR440 pKa = 11.84QPIRR444 pKa = 11.84NSTTAEE450 pKa = 3.97TKK452 pKa = 9.3TAQSKK457 pKa = 10.08QPTPKK462 pKa = 10.03QLSNNTTPEE471 pKa = 3.71NTTPPNNKK479 pKa = 9.6SSSQTDD485 pKa = 3.3AAPTEE490 pKa = 4.38EE491 pKa = 3.95IEE493 pKa = 4.47IRR495 pKa = 11.84SSLWRR500 pKa = 11.84RR501 pKa = 11.84RR502 pKa = 11.84YY503 pKa = 9.84VYY505 pKa = 10.21GPCRR509 pKa = 11.84EE510 pKa = 4.13NVLEE514 pKa = 4.98HH515 pKa = 6.76PMNPCFKK522 pKa = 11.06DD523 pKa = 3.15NTTWIYY529 pKa = 11.09SDD531 pKa = 3.31NGRR534 pKa = 11.84NLPAGYY540 pKa = 10.08YY541 pKa = 10.0DD542 pKa = 3.74SKK544 pKa = 9.64TDD546 pKa = 4.51KK547 pKa = 10.33IICYY551 pKa = 10.15GIYY554 pKa = 9.98RR555 pKa = 11.84GNSYY559 pKa = 10.82CYY561 pKa = 10.74GRR563 pKa = 11.84IEE565 pKa = 4.38CTCKK569 pKa = 10.71NGTGLLSYY577 pKa = 10.6CCNSYY582 pKa = 10.32NWSS585 pKa = 3.4
MM1 pKa = 7.11EE2 pKa = 4.47VKK4 pKa = 10.42VEE6 pKa = 4.08NVGKK10 pKa = 9.88SQEE13 pKa = 4.18LKK15 pKa = 10.47VKK17 pKa = 9.99VKK19 pKa = 10.96NFIKK23 pKa = 10.62RR24 pKa = 11.84SDD26 pKa = 3.73CKK28 pKa = 10.75KK29 pKa = 10.78KK30 pKa = 10.74LFALILGLVSFEE42 pKa = 3.78LTMNIMLSVMYY53 pKa = 10.62VEE55 pKa = 5.09SNEE58 pKa = 3.96ALSLCRR64 pKa = 11.84IQGTPAPRR72 pKa = 11.84DD73 pKa = 3.29NKK75 pKa = 10.64TNTEE79 pKa = 4.0NTKK82 pKa = 10.62KK83 pKa = 9.2EE84 pKa = 4.5TTFHH88 pKa = 5.8TTTTTRR94 pKa = 11.84DD95 pKa = 3.34PEE97 pKa = 4.07VRR99 pKa = 11.84EE100 pKa = 4.23TKK102 pKa = 6.87TTKK105 pKa = 10.36PKK107 pKa = 9.29TNEE110 pKa = 3.85GATSPSRR117 pKa = 11.84NLTTKK122 pKa = 10.91GDD124 pKa = 3.14IHH126 pKa = 5.48QTTRR130 pKa = 11.84ATTEE134 pKa = 3.89AEE136 pKa = 4.05LEE138 pKa = 4.3KK139 pKa = 10.64QSKK142 pKa = 7.06QTIEE146 pKa = 4.46PDD148 pKa = 3.16TSTKK152 pKa = 10.32KK153 pKa = 8.82HH154 pKa = 5.28TPTRR158 pKa = 11.84PSSEE162 pKa = 4.3SPTTTQATAQLTTPTAPKK180 pKa = 10.6ASIAPKK186 pKa = 10.05NRR188 pKa = 11.84QATTKK193 pKa = 9.16KK194 pKa = 8.79TEE196 pKa = 4.02TGTTTTSRR204 pKa = 11.84AKK206 pKa = 9.28KK207 pKa = 8.3TNNPTEE213 pKa = 4.23TATTTLKK220 pKa = 10.3ATTEE224 pKa = 4.02TGKK227 pKa = 10.92GKK229 pKa = 10.31EE230 pKa = 4.45GPTQHH235 pKa = 6.76TIKK238 pKa = 10.2EE239 pKa = 4.32QPEE242 pKa = 4.17TTAGEE247 pKa = 4.39TTTPQSRR254 pKa = 11.84RR255 pKa = 11.84TTSRR259 pKa = 11.84PAPTTKK265 pKa = 10.09TEE267 pKa = 4.14EE268 pKa = 4.02EE269 pKa = 4.35AEE271 pKa = 4.35TTKK274 pKa = 10.42TRR276 pKa = 11.84TTKK279 pKa = 9.28STQTSTGPPGPTRR292 pKa = 11.84STPSKK297 pKa = 9.71TATEE301 pKa = 4.07NNKK304 pKa = 8.77RR305 pKa = 11.84TTTIKK310 pKa = 10.59RR311 pKa = 11.84PNTANTDD318 pKa = 2.94SRR320 pKa = 11.84QQTRR324 pKa = 11.84TTAEE328 pKa = 3.77QDD330 pKa = 3.16RR331 pKa = 11.84QIQTKK336 pKa = 10.31AKK338 pKa = 7.08PTTNGAHH345 pKa = 6.33AQTTTTPEE353 pKa = 4.22HH354 pKa = 5.87NTDD357 pKa = 3.23TTNSTKK363 pKa = 10.44EE364 pKa = 3.74SSKK367 pKa = 10.64EE368 pKa = 3.86DD369 pKa = 3.16KK370 pKa = 8.18TTRR373 pKa = 11.84DD374 pKa = 3.8PSSKK378 pKa = 9.72TPTDD382 pKa = 3.26QEE384 pKa = 4.34DD385 pKa = 3.6ASKK388 pKa = 9.6GTTAANPRR396 pKa = 11.84KK397 pKa = 8.46NTEE400 pKa = 4.0ANTRR404 pKa = 11.84TPPTTTPTRR413 pKa = 11.84HH414 pKa = 4.38TTEE417 pKa = 4.32SATSTTGDD425 pKa = 3.04KK426 pKa = 10.44TKK428 pKa = 11.14AKK430 pKa = 4.72TTRR433 pKa = 11.84WKK435 pKa = 9.48STADD439 pKa = 3.48RR440 pKa = 11.84QPIRR444 pKa = 11.84NSTTAEE450 pKa = 3.97TKK452 pKa = 9.3TAQSKK457 pKa = 10.08QPTPKK462 pKa = 10.03QLSNNTTPEE471 pKa = 3.71NTTPPNNKK479 pKa = 9.6SSSQTDD485 pKa = 3.3AAPTEE490 pKa = 4.38EE491 pKa = 3.95IEE493 pKa = 4.47IRR495 pKa = 11.84SSLWRR500 pKa = 11.84RR501 pKa = 11.84RR502 pKa = 11.84YY503 pKa = 9.84VYY505 pKa = 10.21GPCRR509 pKa = 11.84EE510 pKa = 4.13NVLEE514 pKa = 4.98HH515 pKa = 6.76PMNPCFKK522 pKa = 11.06DD523 pKa = 3.15NTTWIYY529 pKa = 11.09SDD531 pKa = 3.31NGRR534 pKa = 11.84NLPAGYY540 pKa = 10.08YY541 pKa = 10.0DD542 pKa = 3.74SKK544 pKa = 9.64TDD546 pKa = 4.51KK547 pKa = 10.33IICYY551 pKa = 10.15GIYY554 pKa = 9.98RR555 pKa = 11.84GNSYY559 pKa = 10.82CYY561 pKa = 10.74GRR563 pKa = 11.84IEE565 pKa = 4.38CTCKK569 pKa = 10.71NGTGLLSYY577 pKa = 10.6CCNSYY582 pKa = 10.32NWSS585 pKa = 3.4
Molecular weight: 64.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4499 |
71 |
2005 |
499.9 |
55.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.024 ± 1.034 | 1.845 ± 0.281 |
4.445 ± 0.395 | 7.002 ± 0.967 |
3.134 ± 0.725 | 5.446 ± 0.436 |
1.467 ± 0.252 | 5.735 ± 0.521 |
7.691 ± 0.424 | 9.535 ± 1.2 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.489 ± 0.334 | 5.335 ± 0.363 |
4.09 ± 0.556 | 3.401 ± 0.261 |
5.134 ± 0.434 | 8.18 ± 0.377 |
8.402 ± 2.618 | 6.668 ± 1.061 |
1.134 ± 0.251 | 2.845 ± 0.294 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
