
Microcoleus sp. PCC 7113
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Oscillatoriales; Microcoleaceae; Microcoleus; unclassified Microcoleus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
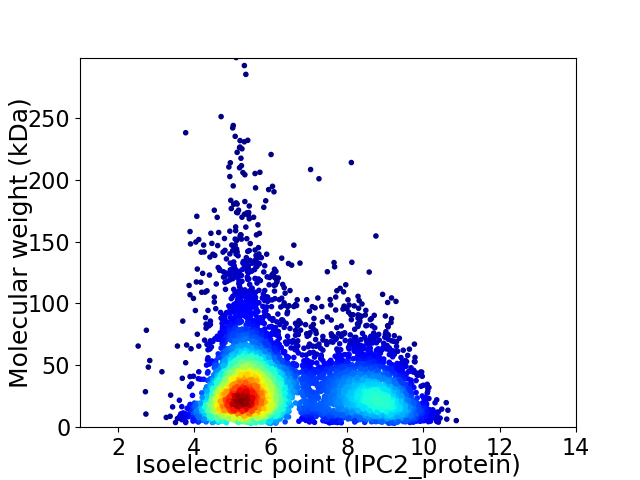
Virtual 2D-PAGE plot for 6397 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9WLA7|K9WLA7_9CYAN PHB domain-containing protein OS=Microcoleus sp. PCC 7113 OX=1173027 GN=Mic7113_5329 PE=4 SV=1
MM1 pKa = 7.15FLEE4 pKa = 4.79TSSEE8 pKa = 4.39TLGSLSSDD16 pKa = 3.47TLKK19 pKa = 10.94HH20 pKa = 6.49LNTAYY25 pKa = 10.79LGLSGSDD32 pKa = 3.38LSSLEE37 pKa = 5.61LSDD40 pKa = 4.36LWGGNIVQGSSLDD53 pKa = 3.92PLSVAASEE61 pKa = 4.54RR62 pKa = 11.84SLEE65 pKa = 3.98TTYY68 pKa = 10.65TIEE71 pKa = 4.1QFNDD75 pKa = 3.29SLIDD79 pKa = 3.65SQTSISGIQGTKK91 pKa = 9.77WNDD94 pKa = 3.2LNGNGVQDD102 pKa = 3.41TGEE105 pKa = 4.31AGLAGWTIYY114 pKa = 10.89LDD116 pKa = 3.56QNNNGTLDD124 pKa = 3.37TGEE127 pKa = 4.34TSTTTDD133 pKa = 2.79ANGNYY138 pKa = 10.12SFTGLAAGTYY148 pKa = 8.13TVAEE152 pKa = 4.48VMQSGWQQTSPGAVTNGSFEE172 pKa = 4.38TGSLTGWNTIGSTEE186 pKa = 4.17VQTASYY192 pKa = 10.57GVTPTQGTYY201 pKa = 9.28QAVLTNGSGSVTDD214 pKa = 3.8SALEE218 pKa = 4.19SFFGWSAGTLDD229 pKa = 4.87GMGNGNATEE238 pKa = 4.38GSAMKK243 pKa = 10.63LSTITVAAGSTLTFDD258 pKa = 3.64WNFLTNEE265 pKa = 4.33STPSSYY271 pKa = 11.49NDD273 pKa = 3.31FAFVSINGLTEE284 pKa = 4.1LADD287 pKa = 3.96TNSTFALSPTVFTEE301 pKa = 3.55QTGYY305 pKa = 11.28KK306 pKa = 9.39KK307 pKa = 10.8FSYY310 pKa = 9.27TFRR313 pKa = 11.84AAGTYY318 pKa = 9.41TIGLGVVDD326 pKa = 5.52AGDD329 pKa = 3.66KK330 pKa = 10.32TVDD333 pKa = 3.23SALVVDD339 pKa = 4.91NFSLTPNGVYY349 pKa = 9.48QVNLASGQTVNNINFGSKK367 pKa = 9.84GVSVLAEE374 pKa = 4.49PNDD377 pKa = 4.47TIPSAIDD384 pKa = 3.28SGLSSSNPGTFKK396 pKa = 11.15DD397 pKa = 3.8KK398 pKa = 11.25GAIGDD403 pKa = 4.05NPNVASDD410 pKa = 4.13LDD412 pKa = 3.67VDD414 pKa = 4.68FVKK417 pKa = 10.68FDD419 pKa = 4.15LKK421 pKa = 11.19AGDD424 pKa = 4.24KK425 pKa = 9.93ATIDD429 pKa = 3.36IDD431 pKa = 4.03TNPLVSSLDD440 pKa = 3.44SFVQVFDD447 pKa = 3.67SAGNLVAFNDD457 pKa = 3.38NGAASGEE464 pKa = 4.3TSSLDD469 pKa = 3.27SFLEE473 pKa = 4.53FTATTTGTYY482 pKa = 10.28YY483 pKa = 10.95LAVSGKK489 pKa = 10.48GNSSYY494 pKa = 11.26NPNVQGSGASGSTGGYY510 pKa = 8.16NLEE513 pKa = 3.99VSVIPKK519 pKa = 9.82FSSNFGYY526 pKa = 11.07GLVDD530 pKa = 3.66AAAAVASATGQDD542 pKa = 3.25SAFADD547 pKa = 3.45VAYY550 pKa = 10.59FGGQNDD556 pKa = 3.42WGVNAVKK563 pKa = 10.48APEE566 pKa = 3.64AWAQGFTGEE575 pKa = 4.56GIVVAVIDD583 pKa = 3.47TGVDD587 pKa = 3.48YY588 pKa = 11.14KK589 pKa = 11.33HH590 pKa = 7.29PDD592 pKa = 3.15LDD594 pKa = 3.9ANIWVNEE601 pKa = 3.92GEE603 pKa = 4.2IAGNGVDD610 pKa = 5.25DD611 pKa = 5.65DD612 pKa = 5.02GNGYY616 pKa = 9.63IDD618 pKa = 6.14DD619 pKa = 3.94VNGWDD624 pKa = 5.35FVAGDD629 pKa = 4.02NDD631 pKa = 3.52AMDD634 pKa = 3.93VYY636 pKa = 11.74GHH638 pKa = 5.85GTHH641 pKa = 6.28VAGTIAAEE649 pKa = 4.18KK650 pKa = 10.64NDD652 pKa = 3.57FGVTGIAYY660 pKa = 7.92NAKK663 pKa = 9.4IMPVKK668 pKa = 10.78VLDD671 pKa = 4.51DD672 pKa = 4.13NGSGSNVDD680 pKa = 3.18VAAGIKK686 pKa = 9.56YY687 pKa = 10.35AADD690 pKa = 3.36NGADD694 pKa = 3.92VINLSLGGGYY704 pKa = 10.11SSEE707 pKa = 3.78IKK709 pKa = 10.66DD710 pKa = 3.47AVEE713 pKa = 3.59YY714 pKa = 8.91ATAKK718 pKa = 9.7GAVVVMAAGNEE729 pKa = 4.43SGSQPGFPASFANQWGVAVGAVDD752 pKa = 4.97SNNKK756 pKa = 8.5MADD759 pKa = 3.64FSNKK763 pKa = 9.93AGSTEE768 pKa = 3.82LDD770 pKa = 3.55YY771 pKa = 11.63VVAPGVDD778 pKa = 3.84VYY780 pKa = 11.22STTPGNTYY788 pKa = 10.5QSFNGTSMATPHH800 pKa = 5.64VAGVAALILSANPNLTPAEE819 pKa = 4.22VEE821 pKa = 4.17SLLTKK826 pKa = 9.15TANPTAITVV835 pKa = 3.4
MM1 pKa = 7.15FLEE4 pKa = 4.79TSSEE8 pKa = 4.39TLGSLSSDD16 pKa = 3.47TLKK19 pKa = 10.94HH20 pKa = 6.49LNTAYY25 pKa = 10.79LGLSGSDD32 pKa = 3.38LSSLEE37 pKa = 5.61LSDD40 pKa = 4.36LWGGNIVQGSSLDD53 pKa = 3.92PLSVAASEE61 pKa = 4.54RR62 pKa = 11.84SLEE65 pKa = 3.98TTYY68 pKa = 10.65TIEE71 pKa = 4.1QFNDD75 pKa = 3.29SLIDD79 pKa = 3.65SQTSISGIQGTKK91 pKa = 9.77WNDD94 pKa = 3.2LNGNGVQDD102 pKa = 3.41TGEE105 pKa = 4.31AGLAGWTIYY114 pKa = 10.89LDD116 pKa = 3.56QNNNGTLDD124 pKa = 3.37TGEE127 pKa = 4.34TSTTTDD133 pKa = 2.79ANGNYY138 pKa = 10.12SFTGLAAGTYY148 pKa = 8.13TVAEE152 pKa = 4.48VMQSGWQQTSPGAVTNGSFEE172 pKa = 4.38TGSLTGWNTIGSTEE186 pKa = 4.17VQTASYY192 pKa = 10.57GVTPTQGTYY201 pKa = 9.28QAVLTNGSGSVTDD214 pKa = 3.8SALEE218 pKa = 4.19SFFGWSAGTLDD229 pKa = 4.87GMGNGNATEE238 pKa = 4.38GSAMKK243 pKa = 10.63LSTITVAAGSTLTFDD258 pKa = 3.64WNFLTNEE265 pKa = 4.33STPSSYY271 pKa = 11.49NDD273 pKa = 3.31FAFVSINGLTEE284 pKa = 4.1LADD287 pKa = 3.96TNSTFALSPTVFTEE301 pKa = 3.55QTGYY305 pKa = 11.28KK306 pKa = 9.39KK307 pKa = 10.8FSYY310 pKa = 9.27TFRR313 pKa = 11.84AAGTYY318 pKa = 9.41TIGLGVVDD326 pKa = 5.52AGDD329 pKa = 3.66KK330 pKa = 10.32TVDD333 pKa = 3.23SALVVDD339 pKa = 4.91NFSLTPNGVYY349 pKa = 9.48QVNLASGQTVNNINFGSKK367 pKa = 9.84GVSVLAEE374 pKa = 4.49PNDD377 pKa = 4.47TIPSAIDD384 pKa = 3.28SGLSSSNPGTFKK396 pKa = 11.15DD397 pKa = 3.8KK398 pKa = 11.25GAIGDD403 pKa = 4.05NPNVASDD410 pKa = 4.13LDD412 pKa = 3.67VDD414 pKa = 4.68FVKK417 pKa = 10.68FDD419 pKa = 4.15LKK421 pKa = 11.19AGDD424 pKa = 4.24KK425 pKa = 9.93ATIDD429 pKa = 3.36IDD431 pKa = 4.03TNPLVSSLDD440 pKa = 3.44SFVQVFDD447 pKa = 3.67SAGNLVAFNDD457 pKa = 3.38NGAASGEE464 pKa = 4.3TSSLDD469 pKa = 3.27SFLEE473 pKa = 4.53FTATTTGTYY482 pKa = 10.28YY483 pKa = 10.95LAVSGKK489 pKa = 10.48GNSSYY494 pKa = 11.26NPNVQGSGASGSTGGYY510 pKa = 8.16NLEE513 pKa = 3.99VSVIPKK519 pKa = 9.82FSSNFGYY526 pKa = 11.07GLVDD530 pKa = 3.66AAAAVASATGQDD542 pKa = 3.25SAFADD547 pKa = 3.45VAYY550 pKa = 10.59FGGQNDD556 pKa = 3.42WGVNAVKK563 pKa = 10.48APEE566 pKa = 3.64AWAQGFTGEE575 pKa = 4.56GIVVAVIDD583 pKa = 3.47TGVDD587 pKa = 3.48YY588 pKa = 11.14KK589 pKa = 11.33HH590 pKa = 7.29PDD592 pKa = 3.15LDD594 pKa = 3.9ANIWVNEE601 pKa = 3.92GEE603 pKa = 4.2IAGNGVDD610 pKa = 5.25DD611 pKa = 5.65DD612 pKa = 5.02GNGYY616 pKa = 9.63IDD618 pKa = 6.14DD619 pKa = 3.94VNGWDD624 pKa = 5.35FVAGDD629 pKa = 4.02NDD631 pKa = 3.52AMDD634 pKa = 3.93VYY636 pKa = 11.74GHH638 pKa = 5.85GTHH641 pKa = 6.28VAGTIAAEE649 pKa = 4.18KK650 pKa = 10.64NDD652 pKa = 3.57FGVTGIAYY660 pKa = 7.92NAKK663 pKa = 9.4IMPVKK668 pKa = 10.78VLDD671 pKa = 4.51DD672 pKa = 4.13NGSGSNVDD680 pKa = 3.18VAAGIKK686 pKa = 9.56YY687 pKa = 10.35AADD690 pKa = 3.36NGADD694 pKa = 3.92VINLSLGGGYY704 pKa = 10.11SSEE707 pKa = 3.78IKK709 pKa = 10.66DD710 pKa = 3.47AVEE713 pKa = 3.59YY714 pKa = 8.91ATAKK718 pKa = 9.7GAVVVMAAGNEE729 pKa = 4.43SGSQPGFPASFANQWGVAVGAVDD752 pKa = 4.97SNNKK756 pKa = 8.5MADD759 pKa = 3.64FSNKK763 pKa = 9.93AGSTEE768 pKa = 3.82LDD770 pKa = 3.55YY771 pKa = 11.63VVAPGVDD778 pKa = 3.84VYY780 pKa = 11.22STTPGNTYY788 pKa = 10.5QSFNGTSMATPHH800 pKa = 5.64VAGVAALILSANPNLTPAEE819 pKa = 4.22VEE821 pKa = 4.17SLLTKK826 pKa = 9.15TANPTAITVV835 pKa = 3.4
Molecular weight: 85.73 kDa
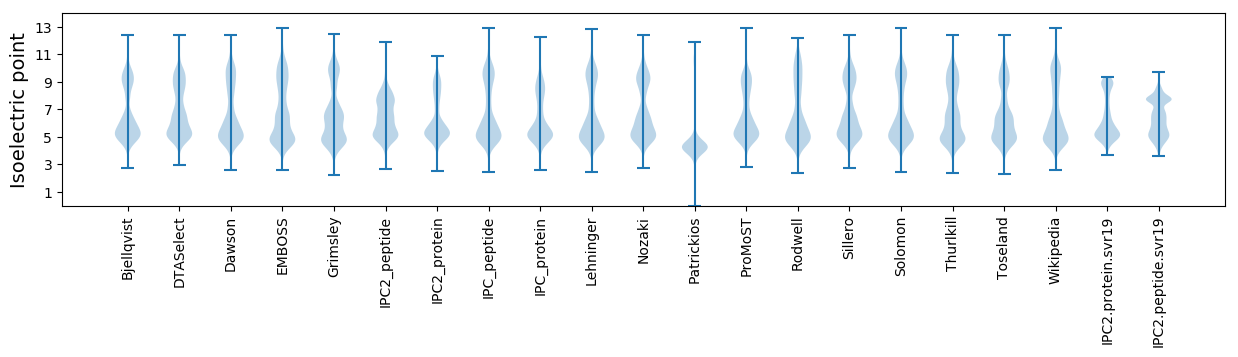
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9WEJ7|K9WEJ7_9CYAN NB-ARC domain-containing protein OS=Microcoleus sp. PCC 7113 OX=1173027 GN=Mic7113_2380 PE=4 SV=1
MM1 pKa = 7.54GLPTRR6 pKa = 11.84GEE8 pKa = 4.21ARR10 pKa = 11.84NLAYY14 pKa = 9.53MKK16 pKa = 10.95LMMIGWLGIQGTASALPVKK35 pKa = 10.02PLALPPSPPLLTAQSPASPPSSKK58 pKa = 10.33CPRR61 pKa = 11.84DD62 pKa = 3.65VEE64 pKa = 4.59TLTSLMLKK72 pKa = 10.17DD73 pKa = 3.88LPSYY77 pKa = 10.77ANRR80 pKa = 11.84VMQRR84 pKa = 11.84SRR86 pKa = 11.84RR87 pKa = 11.84SVRR90 pKa = 11.84TPSSSSSVLVAGRR103 pKa = 11.84PEE105 pKa = 4.22FEE107 pKa = 4.91PLTLGPGSYY116 pKa = 9.34TPPASAIDD124 pKa = 4.2LEE126 pKa = 4.76PPQQVFFTTLEE137 pKa = 3.87RR138 pKa = 11.84QYY140 pKa = 11.47LGGKK144 pKa = 8.99ASYY147 pKa = 8.27MQLYY151 pKa = 9.09HH152 pKa = 7.0WLFLTQTPQGWRR164 pKa = 11.84VATMFSRR171 pKa = 11.84IGSSSGRR178 pKa = 11.84PPTPPQEE185 pKa = 4.15SSKK188 pKa = 10.96GVVGQAVNIWLRR200 pKa = 11.84DD201 pKa = 3.6CRR203 pKa = 11.84AGAIRR208 pKa = 11.84QSRR211 pKa = 11.84PRR213 pKa = 11.84NQKK216 pKa = 9.68
MM1 pKa = 7.54GLPTRR6 pKa = 11.84GEE8 pKa = 4.21ARR10 pKa = 11.84NLAYY14 pKa = 9.53MKK16 pKa = 10.95LMMIGWLGIQGTASALPVKK35 pKa = 10.02PLALPPSPPLLTAQSPASPPSSKK58 pKa = 10.33CPRR61 pKa = 11.84DD62 pKa = 3.65VEE64 pKa = 4.59TLTSLMLKK72 pKa = 10.17DD73 pKa = 3.88LPSYY77 pKa = 10.77ANRR80 pKa = 11.84VMQRR84 pKa = 11.84SRR86 pKa = 11.84RR87 pKa = 11.84SVRR90 pKa = 11.84TPSSSSSVLVAGRR103 pKa = 11.84PEE105 pKa = 4.22FEE107 pKa = 4.91PLTLGPGSYY116 pKa = 9.34TPPASAIDD124 pKa = 4.2LEE126 pKa = 4.76PPQQVFFTTLEE137 pKa = 3.87RR138 pKa = 11.84QYY140 pKa = 11.47LGGKK144 pKa = 8.99ASYY147 pKa = 8.27MQLYY151 pKa = 9.09HH152 pKa = 7.0WLFLTQTPQGWRR164 pKa = 11.84VATMFSRR171 pKa = 11.84IGSSSGRR178 pKa = 11.84PPTPPQEE185 pKa = 4.15SSKK188 pKa = 10.96GVVGQAVNIWLRR200 pKa = 11.84DD201 pKa = 3.6CRR203 pKa = 11.84AGAIRR208 pKa = 11.84QSRR211 pKa = 11.84PRR213 pKa = 11.84NQKK216 pKa = 9.68
Molecular weight: 23.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2155269 |
29 |
2656 |
336.9 |
37.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.262 ± 0.032 | 0.984 ± 0.011 |
4.817 ± 0.023 | 6.43 ± 0.039 |
3.776 ± 0.02 | 6.773 ± 0.041 |
1.857 ± 0.018 | 6.129 ± 0.026 |
4.476 ± 0.032 | 11.161 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.875 ± 0.014 | 4.016 ± 0.029 |
4.997 ± 0.03 | 5.642 ± 0.031 |
5.456 ± 0.028 | 6.611 ± 0.029 |
5.73 ± 0.03 | 6.666 ± 0.024 |
1.461 ± 0.015 | 2.88 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
